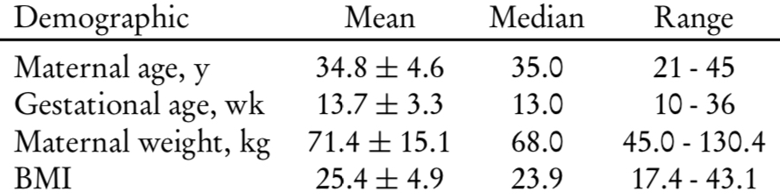Research Article
Volume 2 Issue 2 - 2020
Performances of a Snp-Based Noninvasive Prenatal Test
1Center for Gynecology and Obstetrics, University Hospital Gießen & Marburg, Department for Prenatal Medicine, Giessen / DE, Germany
2Clinic for Gynecology and Obstetrics, University Hospital RWTH Aachen, Department for Prenatal Medicine, Aa-chen / DE, Germany
2Clinic for Gynecology and Obstetrics, University Hospital RWTH Aachen, Department for Prenatal Medicine, Aa-chen / DE, Germany
*Corresponding Author: Malena Götte, Center for Gynecology and Obstetrics, University Hospital Gießen & Marburg, Department for Prenatal Medi-cine, Giessen/DE, Germany.
Received: July 13, 2020; Published: July 22, 2020
Abstract
Noninvasive prenatal tests based on cell free DNA have spread broadly, with many studies regarding their performance and improvement being published in the last few years. Their use in the clinical practice, in most countries, is not included in the common screening and must be paid by the patient. In this study we analyzed the performance of a noninvasive test based on single nucleotide polymorphisms (Panorama® by Natera) on patients handled in the prenatal diagnostic unit of the University Hospital of Giessen and Marburg in Giessen (Germany) from July 2016 to July 2018. We analyzed the results of the tests in 440 patients, in a mixed population of high-risk and low-risk women. For "high risk" and "no call" results in the cell free DNA analysis, we were able to collect follow-up information. For the validation of our high-risk results we considered informations obtained from a prenatal karyotype or from the clinical evaluation at birth and postpartal episode. 426/440 patients received a result, with 16 High Risk results and 410 Low Risk results. The combined high risk rate for all indications was 3.75% (16/426), with 11 cases of trisomy 21, 2 of trisomy 18, 1 of trisomy 13 and 2 of sex chromosome aneuploidies (XXY and XXX). The total no call rate was 23/460 submitted tests (5.00%). The main reason for the no call results was a low fetal fraction (< 3.5%). The total rate of aneuploidies in the population with a call is 2.91% (12/412, patients with follow up informations). The aneuploidy rate in no call results is 31,6% and is thus much higher. This difference is statistically significative (p-value <0.0001). The calculated test performance shown as positive predictive value for trisomy 21 was 90.9% and for all four aneuploidies combined was 92.9%.
In Conclusion, the performance of Panorama® (calculated with positive predictive value) proved much better than that of standard screening (First trimester scan) and is comparable to the NIPT test performance described in the literature. In addition, a no call result might be linked with a higher risk of aneuploidies.
Keywords: Fetal Medicine, Ultrasound; NIPT; Noninvasive Prenatal Test; SNP
Nicht invasive Pränataltests werden heutzutage häufig in der Klinik eingesetzt. Ihre Möglichkeiten ,sowie die Testgüte wurden bereits durch viele Studien analysiert und bestätigt. In den meisten Ländern ist der Einsatz von nicht investiven Pränataltests keine Leistung der Krankenkassen und die Patienten müssen die Kosten selbst tragen. In dieser Studie haben wir die Testperfomance des Panorama®-Tests untersucht. Es wurden insgesamt 440 Patientinnen inkludiert, die im Zeitraum von Juli 2016 bis Juli 2018 in der Abteilung für Pränataldiagnostik am UKGM in Gießen vorgestellt wurden. Das Patientinnenkollektiv besteht aus einer Mischpopulation mit Hoch- und Niedrigrisiko Patientinnen. Bei auffälligen Testergebnissen erfolgte die detaillierte Follow-Up-Evaluation und größtenteils auch die Karyotypisierung mittels invasiver Diagnostik. 426/440 Patientinnen erhielten ein Testergebnis, mit insgesamt 16 High Risk Ergebnissen und 410 Low Risk Ergebnissen. Die kombinierte High Risk Rate betrug 3,75% (16/426), mit 11 Fällen von Trisomie 21, zwei Fällen Trisomie 18, einem Trisomie 13 Fall und zwei Fällen mit geschlechtschromosomalen Störungen (XXY und XXX). Die totale No-Call-Rate mit 23/460 Tests beträgt 5%. Der Hauptgrund für ein No-Call Ergebnis ist eine zu niedrige fetale Fraktion (<3,5%). Die totale Rate an Aneuploidien in unserer Population liegt bei 2,91% (12/412, Patientinnen mit Follow-Up Daten). Die Aneuploidie-Rate in der No-Call Kohorte liegt mit 31,6% signifikant höher (p-value <0.0001). Die Testperfomance für Trisomie 21 ergibt einen positiven prädikativen Wert von 90,9% und kombiniert für alle vier Aneuploidien einen Wert von 92,9%.
Zusammenfassend kann man sagen, dass die Testperfomance des Panorama®-Tests, hier bestimmt über den positiven prädikativen Wert, bessere Werte als das Standardscreening (Ersttrimesterscreening) aufweist und die Testperformance vergleichbar mit den Werten aus der Literatur ist. Zusätzlich zeigen unsere Daten, dass ein No-Call Ergebnis mit einem höheren Aneuploidierisiko assoziiert ist.
Introduction
Every woman has a risk to carry a fetus with a chromosomal anomaly. Prenatal screening is aimed to determine the individual risk of the pregnancy, in order to offer invasive diagnostic only in high-risk pregnancies. In the early 1970’s, the high risk group was selected only on the basis of maternal age, thus pregnant women older than 40 years were offered amniocentesis. This group was later extended to women older than 35 years as the application of amni-ocentesis spread and its safety increased. Today, to calculate the individual risk, a background risk (or a priori risk) is multiplied by a series of factors (or likelihood ratios). The latter depend on biochemical tests and sonographic findings. The background risk depends on maternal and gestational age. The risk of trisomies increases with mater-nal age, but it decreases with gestational age, as many chromosomal abnormalities are associated with a high in utero mortality. Previously affected pregnancies also represent a risk factor, as in a small fraction of cases chromo-somal anomalies are caused by a parental mosaicism or genetic defect that lead to nondisjunction. Other risk factors are obtained by testing biochemical values and sonographic features that are more frequent in chromosomally ab-normal than in normal fetuses. The likelihood ratio for every measurement is calculated by dividing the percentage of affected fetuses by the percentage of normal fetuses with that measurement [1].
In 1997 Lo et al. proved the presence of cell free DNA (cfDNA) derived from the Y chromosome in both the plasma and the serum of pregnant women carrying male fetuses [2]. Following the discovery of cfDNA, many researchers attempted to develop non-invasive prenatal tests (NIPTs) based upon it. Initially, it was used to determine the fetal sex and, in RhD negative women, the RhD status of the fetus, to avoid fetal hemolytic disease [3]. The next step was the development of a test to detect chromosomal aneuploidies, and the first screening test for trisomies based on cfDNA was released on the market in 2011 in the USA [4]. Later, several European clinical research sites took part in a large NIPT study, by Norton et al., in which the samples were sent to the Ariosa, Inc. laboratory in the USA to be analyzed [5]. NIPTs based on cfDNA are now broadly used in prenatal diagnostic. They can recognize the most common aneuploidies, micro deletions and micro duplication in the fetal genome, and determine the fetal sex. While testing, the proportion of cfDNA in the maternal plasma is referred to as the fetal fraction (FF) and is expressed in percentage of the total cfDNA. It is detectable as early as 4 weeks of gestation and it consists of fragments of approx-imately 150 base pairs that represent the whole fetal genome [6]. In singleton pregnancies the FF is normally among 3-13% in the 10th pregnancy week, and it increases gradually of 0.1% per week up to the 21st week, and afterwards of 1% per week [7]. The FF depends mainly on the placental volume, maternal weight, gestational age, singleton or twin-pregnancies and partly on chromosomal abnormalities caused by a small placenta [8-9]. Today the specifica-tion of the FF in NIPTs is mandatory.
Material and Methods
In this analysis we used the Panorama®-Test, based on the analysis of SNPs (single nucleotide polymorphisms). SNPs are the most common type of genetic variation among individuals. The amplification through polymerase chain reaction (PCR) and the sequencing of more than 13000 SNP allows the distinction between the maternal and fetal cfDNA. This also enables the quantification of their relative contributions to the total cfDNA, and thus the assessment of FF. The performance of this method, like for the other techniques, is affected by a low FF. Maternal genotype is obtained by leukocytes present in the blood sample, and this allows to identify the specific SNPs of the mother [7]. The sequencing products are then evaluated based on the hypothesis that the fetus is euploid, aneuploid (trisomic or monosomic) and triploid. Then, a Bayesian-based maximum likelihood for the chromosomal status of the fetus is calculated, taking also into account that there could have been recombination. An excess (or deficiency) of a chromosome copy is identified as an increased number of fetal SNPs located on that chromosome. As a consequence, this is the only method that allows to detect triploidy, as it does not require a reference chromosome [4]. This method can also identify consanguinity, non-paternity, uniparental disomy and parental origin of aneuploidy, recombination and inherited mutations [6]. It is also possible to recognize the influence of a Vanishing Twin, but not possible to give a valid result. From March 2014, screening for clinically significant micro deletion syndromes with the SNP-based method is offered. From October 2017 on, screening for aneuploidies is possible also for twin pregnancies, egg donors and surrogates [10].
Our study population included high-risk and low-risk patients. Two 10 ml Streck-Tubes were collected from each patient. From July 2016 to September 2017 the samples were sent to Natera Laboratories in the USA to be analyzed. From September 2017 on, the analysis was performed by Zotz-Klimas laboratory in Düsseldorf. The analysis was performed on chromosomes 21, 18, 13, X and Y for all samples. Trisomy 21, 18, 13 and Monosomy X were screened in all patients. Sex chromosome anomalies and triploidy were also reported when present. Patients could also choose additional screening for five microdeletion syndromes (22q11.2, 1p36, cri-du-chat, Prader-Willi and Angelman Syndrome), but had to pay higher costs. Initially twin pregnancies and pregnancies based on egg donation were considered "out of specification" and the analysis could not be performed. Since October 2017 the screening for trisomies was also possible in this situations. In the case of a monozygotic twin pregnancy, the test could screen monosomy X, sex chromosome aneuploidies and 22q11.2 deletion as well. Patients in our clinic firstly got a detailed anomaly ultrasound and genetic counselling. Afterwards the patients decided between no further testing, conventional FTS (between 11+0 and 13+6 weeks) and/or NIPT. We included in our study the group of patients that opted for NIPT.
The report includes a risk score for each aneuploidy (and potentially for micro duplication and microdeletion syndromes) and, when requested, the fetal sex. The cutoff for high risk results was a risk score ≥ 1/100, while samples with a risk score <1/100 were considered low risk. To determine the performance of NIPT tests, we classified each sample based on the confirmed outcome. We divided the samples in true positives (TP), false positives (FP) and false negatives (FN) on the basis of confirmative procedures or clinical evaluation after birth. Samples that did not have available information about the outcome were labeled as "without follow-up". One limit of clinical retrospective studies is the lack of follow-up information about negative (low risk) results. Therefore, it was not possible to identify true negatives (TN), and FP were expected to be voluntarily reported by patients, or were highlighted by a discordance in the ultrasound examination. The most used parameters to express a test performance are sensitivity and specificity. Because of the lack of information about negative results, we could not determine these parameters. We calculated instead the positive predictive value (PPV), which is the ratio of true positives to all positive results [TP/ (TP+FP)].
Results
In the period considered 440 Patients choose NIPT. The mean maternal age was 34.8 years, with 57.9% of women with an age ≥35 years. Patient’s demographics are detailed in Table 1.
Mean gestational age at the first NIPT was 13.7 weeks, with 66.8% of samples drawn in the first trimester and 32.5% in the second trimester. If we take the redraws after a "no call" result, into the account, the samples drawn in the second trimester are increasing to 35% of the total (Table 2).
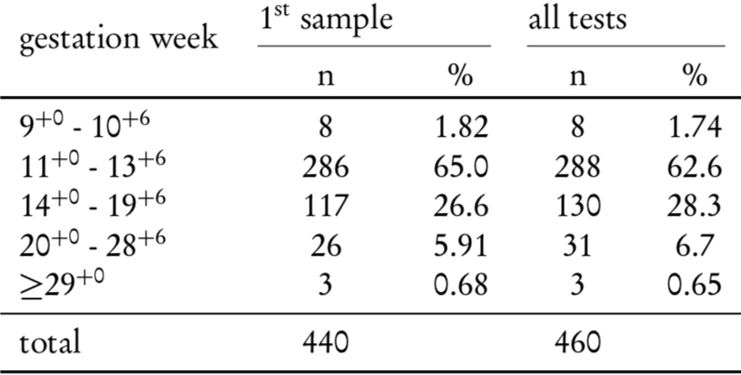
Table 2: Number of samples stratified for gestational week. Redraws are taken into account in the third column.
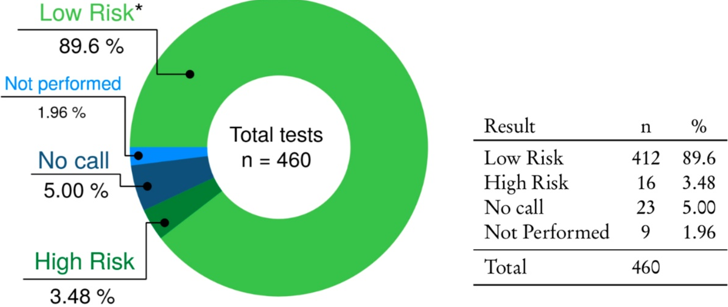
The total number of NIPT performed in the period considered is 460. Figure 1 shows the proportion of high risk, low risk and "no call" results. In Figure 2 you can see the flowchart of our study.
Of the 440 sample submitted, 9 were not analyzed (not performed). Causes of not performed samples are for exam-ple caused by a delay of the transportation or caused by a twin- or an egg-donor sample (tested before October 2017). For the 9 samples that were not analyzed, five (55.5%) submitted a second sample for the analysis, two choose a NIPT test based on the counting method and two didn’t submit a new sample.
Calls
426/440 of our patients received a result, with 16 High Risk results and 410 Low Risk results (Figure 3). Actually, three of the 410 low risk samples resulted low risk for four of the five chromosomes considered, and no call for chromosome X. These are herein considered as low risk. One of these cases resulted normal in the confirmatory diagnostic, while the other two patients did not want any further investigation. The combined high risk rate for all indications was 3.75% (16/426), with in total 11 cases of trisomy 21, two of trisomy 18, one of trisomy 13 and two cases of sex chromosome aneuploidies (XXY and XXX). One high risk result for trisomy 21 was classified as false positive because the karyotype, obtained from a confirmatory invasive procedure, was normal. The two samples at high risk for sex chromosome aneuploidies did not have any follow-up.
426/440 of our patients received a result, with 16 High Risk results and 410 Low Risk results (Figure 3). Actually, three of the 410 low risk samples resulted low risk for four of the five chromosomes considered, and no call for chromosome X. These are herein considered as low risk. One of these cases resulted normal in the confirmatory diagnostic, while the other two patients did not want any further investigation. The combined high risk rate for all indications was 3.75% (16/426), with in total 11 cases of trisomy 21, two of trisomy 18, one of trisomy 13 and two cases of sex chromosome aneuploidies (XXY and XXX). One high risk result for trisomy 21 was classified as false positive because the karyotype, obtained from a confirmatory invasive procedure, was normal. The two samples at high risk for sex chromosome aneuploidies did not have any follow-up.
No-Calls
Of the 436 analyzed samples, 19 resulted in a no call for all the tested conditions. The main reason for a no call results was a low fetal fraction (< 3.5%), followed by specific situations that caused problems in the analysis (e.g.patient with liver transplant). 12 out of 19 patients receiving a no call result submitted a second sample (63.1%). Nine of them received a risk call, with one high risk result. Among the remaining patients, four choose to undergo amniocentesis, one performed a NIPT test from another company and two refused any further investigation.
Of the 436 analyzed samples, 19 resulted in a no call for all the tested conditions. The main reason for a no call results was a low fetal fraction (< 3.5%), followed by specific situations that caused problems in the analysis (e.g.patient with liver transplant). 12 out of 19 patients receiving a no call result submitted a second sample (63.1%). Nine of them received a risk call, with one high risk result. Among the remaining patients, four choose to undergo amniocentesis, one performed a NIPT test from another company and two refused any further investigation.
Additionally, we had a single case of sex discordance in a low risk report. The fetus was classified as male in the NIPT test, but was defined as female in the ultrasound. A second sample for NIPT testing was then submitted for sex determination, and it returned as a no call result. A following amniocentesis revealed a Turner mosaic (45, X0/46, XY 20%). The first NIPT result was therefore classified as false negative. Overall, the total no callrate was 5% (23 tests on a total of 460 tests submitted).
Follow-up information were available for 19 of 20 patients that received at least one no call result, and six of them (31.6%) had a confirmed aneuploidy. If you take a further look on these aneuploid cases, it is remarkable that five of these six cases showed sonographic abnormalities before. The other case had a pathological first trimester scan. The patients have been counselled in detail, but did not want an invasive procedure and choose to make a NIPT first and. The rate of aneuploidies in the population with a call (i.e. patients receiving a result in the first sample) with follow up is 2.91% (12/412). The aneuploidy rate in no call results is 31.6% and accordingly much higher (Figure 4). As previously stated by Pergament et al.[11], the difference is statistically significative (p-value <0.0001). This translates into an odd ratio of 15 (C.I. 5.0 - 47.4). The reason behind this is probably the link between aneuploidies and a small placenta that result in a low fetal fraction (in our study the major cause of no callresults). This shows that in cases with no result in NIPT, special attention should be paid and the higher risk of aneuploidy should be included into counselling. Ideally, this problem should be addressed in the pre-test counselling, and patients should be informed of the possibility of no callresult and its implications.
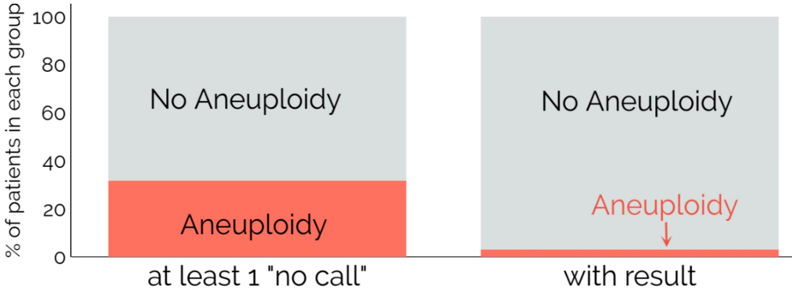
Figure 4: The aneuploidy rate among patients receiving at least one No-Call result is significantly higher than in the population with a call.
Performance
To assess the performance of the test, we considered only cases with confirmed outcome. A limitation in the performance, which is the main issue of many clinical studies on the subject, is the lack of follow-up regarding low risk results. In this study the false negative result was highlighted by the discordance with ultrasound examination. As previously mentioned, low risk cases lack follow-up information, not allowing to assess the specificity and sensitivity of the test. Therefore, we calculated the Positive Predictive Value (PPV), the detection rate (DR) and the false positive rate (FPR) on the basis of cases with a confirmed outcome. The PPV values for the tested conditions are shown in Table 3. The PPV for trisomy 21 was 90.9% and the PPV for all four aneuploidies combined is 92.9%. This PPV values are in line with those shown in previous studies for NIPT tests based on SNPs: Dar, et al.reported a PPV of 90.9% for Trisomy 21 and of 82.9% for all aneuploidies combined [12]. Eiben, et al.reported a PPV of 97.4% for trisomy 21 and 90.8% for the combined aneuploidies [13]. This exceeds by far the PPV of standard screening (FTS) for all aneuploidies combined, which is reported by various studies as between 4.7% and 9.20% [14-17]. The positive predictive values for trisomy 21 are instead reported ranging between 4.05 and 4.9% [16-17]. In a study dated 2014, Bianchi et al.already compared screening based on cfDNA and standard screening, showing that the PPV value of NIPT for trisomy 21 was 10-fold than that of combined test (therein reported as 4.2%) [18].
To assess the performance of the test, we considered only cases with confirmed outcome. A limitation in the performance, which is the main issue of many clinical studies on the subject, is the lack of follow-up regarding low risk results. In this study the false negative result was highlighted by the discordance with ultrasound examination. As previously mentioned, low risk cases lack follow-up information, not allowing to assess the specificity and sensitivity of the test. Therefore, we calculated the Positive Predictive Value (PPV), the detection rate (DR) and the false positive rate (FPR) on the basis of cases with a confirmed outcome. The PPV values for the tested conditions are shown in Table 3. The PPV for trisomy 21 was 90.9% and the PPV for all four aneuploidies combined is 92.9%. This PPV values are in line with those shown in previous studies for NIPT tests based on SNPs: Dar, et al.reported a PPV of 90.9% for Trisomy 21 and of 82.9% for all aneuploidies combined [12]. Eiben, et al.reported a PPV of 97.4% for trisomy 21 and 90.8% for the combined aneuploidies [13]. This exceeds by far the PPV of standard screening (FTS) for all aneuploidies combined, which is reported by various studies as between 4.7% and 9.20% [14-17]. The positive predictive values for trisomy 21 are instead reported ranging between 4.05 and 4.9% [16-17]. In a study dated 2014, Bianchi et al.already compared screening based on cfDNA and standard screening, showing that the PPV value of NIPT for trisomy 21 was 10-fold than that of combined test (therein reported as 4.2%) [18].

Table 2: Number of samples stratified for gestational week. Redraws are taken into account in the third column.
Discussion
This study`s main goal was to investigate the performance of NIPT in daily routine. The NIPT considered in this study is Panorama® by Natera. We studied its performance in a low-risk and high-risk population in the prenatal diagnosis unit of the University Hospital of Gießen&Marburg. The study was conducted on the results of samples collected on 440 patients undergoing NIPT. High risk and no call result were validated on the basis of a confirmatory karyotype obtained with invasive procedures or of a clinical evaluation after birth. The analysis of results demon-strates that the positive predictive value (PPV) of NIPT in the clinical practice (92.9%) is much higher than the PPV of standard screening, i.e. combined screening in the first trimester (4.7% - 9.2%) [14-17].
This was previously highlighted by other studies [18], and demonstrates that the performance of tests based on cfDNA in the clinical practice is much superior to that of standard screening. The only obstacle on this path towards NIPTs is their costs, but not their effectiveness. But to mention some problems of the test, the most problematic aspect of Panorama are No-Calls, that are mostly caused by a low FF. Some studies suggest that the re-test after a No-Call result should be performed by a test based on counting methods (wich seem to be more precise in case of low fetal fraction) [19]. At least, the number of no calls decreased in the period of our study, because the cut off of the FF improved. Im comparison to the literature, the No-Call rate in total is similar to previous results [20]. Today, the cut-off concerning the FF depends on the different NIPT-tests and not on the respective method. The values of the fetal fraction are mostly between 2.8% and 4 %. It is however important to always report fetal fraction, to correctly evaluate the results. Furthermore, it is important to note that a no call result it is not a neutral result, as the probabil-ity of an anomaly is higher when the FF is low. In this study we highlighted that 6 out of 19 (31.6%) follow-up pa-tients with a no call results had a confirmed aneuploidy. The rate of aneuploidy in patients with a call (high- and low-risk) was only 12/412 (2.91%). This translates into an odd ratio of 15 (C.I. 5.0 - 47.4), meaning that samples with a no call result were 15 times more likely to have an aneuploidy. An explanation for this can be that low FF is sometimes the consequence of a small placenta, which is a defect often linked with aneuploidies. But is is remarka-ble, that every of this cases showed abnormalities in the ultrasound or a pathological FTS and this fact should be mentioned in the counseling of the patients and the use of NIPTs.
Conclusion
To conclude, the results of our study show a higher PPV for aneuploidies than the standard screening (FTS) and the test performance of the used NIPTs in our setting is comparable to the published data. But all in all it should be used wisely in daily routine and its use should encounter a detailed anomaly ultrasound and thorough genetic counselling.
References
- K.H.Nicolaides. (2003). Screeningforchromosomaldefects. Ultrasound in Obstetrics and Gynecology, vol. 21, , pp. 313–321.
- Y. M. D. Lo, N. Corbetta, P. F. Chamberlain, V. Rai, I. L. Sargent, C. W. Redman, and J. S. Wainscoat. (1997). Presence of fetal DNA in maternal plasma and serum, The Lancet, vol. 350, pp. 485– 487.
- T. Goldwaser and S. Klugman. (2018). Cell-free DNA for the detection of fetal aneuploidy, Fertility and Sterility, vol. 109, pp. 195–200.
- P. Benn. (2014). Non-Invasive Prenatal Testing Using Cell Free DNA in Maternal Plasma: Recent De- velopments and Future Prospects, Journal of Clinical Medicine, vol. 3, pp. 537–565.
- E. J. Verweij, B. Jacobsson, P. A. van Scheltema, M. A. de Boer, M. J. V. Hoffer, D. Hollemon, M. Westgren, K. Song, and D. Oepkes. (2013). European Non-Invasive Trisomy Evaluation (EU-NITE) study: a multicenter prospective cohort study for non-invasive fetal trisomy 21 testing, Prenatal Diagnosis, pp. 1–6.
- P. Benn, H. Cuckle, and E. Pergament. (2013). Non-invasive prenatal testing for aneuploidy: current status and future prospects, Ultrasound in Obstetrics & Gynecology, vol. 42, pp. 15–33.
P.Kozlowski. (2016). NichtinvasivepränataleTests, Der Gynäkologe, vol.49, pp. 415–421. - G. Ashoor, L. Poon, A. Syngelaki, B. Mosimann, and K. H. Nicolaides. (2012). Fetal Fraction in Ma- ternal Plasma Cell-Free DNA at 11-13 Weeks’ Gestation: Effect of Maternal and Fetal Factors, Fetal Diagnosis and Therapy, vol. 31 no. 4, pp. 237–243.
- C. A. Struble, A. Syngelaki, A. Oliphant, K. Song, and K. H. Nicolaides. (2014). Fetal Fraction Esti- mate in Twin Pregnancies Using Directed Cell-Free DNA Analysis, Fetal Diagnosis and Therapy, vol. 35, no. 3, pp. 199–203.
- Natera’sPanoramaNon-InvasivePrenatalTestNowAvailableforScreeningTwinPregnancies,” www.natera.com, 2017.
- E. Pergament, H. Cuckle, B. Zimmermann, M. Banjevic, S. Sigurjonsson, A. Ryan, M. P. Hall, M. Dodd, P. Lacroute, M. Stosic, N. Chopra, N. Hunkapiller, D. E. Prosen, S. McAdoo, Z. Demko, A. Siddiqui, M. Hill, and M. Rabinowitz. (2014). Single-nucleotide polymorphism-based noninvasive prenatal screening in a high-risk and low-risk cohort, Obstetrics and Gynecology, vol. 124, pp. 210–218.
- P. Dar, K. J. Curnow, S. J. Gross, M. P. Hall, M. Stosic, Z. Demko, B. Zimmermann, M. Hill, S. Sigurjonsson, A. Ryan, M. Banjevic, P. L. Kolacki, S. W. Koch, C. M. Strom, M. Rabinowitz, and P. Benn. (2014). Clinical experience and follow-up with large scale single-nucleotide polymorphism- based noninvasive prenatal aneuploidy testing, American Journal of Obstetrics and Gynecology, vol. 211, pp. 1–527.
- B.Eiben, M.Krapp, H.Borth, N.Kutur, P.Kreiselmaier, R.Glaubitz, J.Deutinger, E.Merz. (2015). Single Nucleotide Polymorphism-Based Analysis of Cell-Free Fetal DNA in 3000 Cases from Germany and Austria, Ultrasound International Open, vol. 01, pp. E8–E11.
- B.Eiben, R.Glaubitz, K.O.Kagan. (2014). NichtinvasivePränataldiagnostik, medizinischegenetik, vol. 26, no. 4, pp. 382–390.
- C. Hörmansdörfer, P. Schmidt, P. Hillemanns, and A. Scharf. (2007). Die pränatale Detektion der Tri- somien 13, 18 und 21: Vergleich des Advanced First Trimester Screenings (AFS)?R mit dem Ersttrimester-Screening nach Nicolaides, Zeitschrift für Geburtshilfe und Neonatologie, vol. 211, pp. 243–249.
- J. E. Siljee, A. C. Knegt, M. F. C. M. Knapen, M. N. Bekker, G. H. A. Visser, and P. C. J. I. Schielen. (2014). Positive predictive values for detection of trisomies 21, 18 and 13 and termination of pregnancy rates after referral for advanced maternal age, first trimester combined test or ultra- sound abnormalities in a national screening programme (2007-2009), Prenatal Diagnosis, vol. 34, pp. 259–264.
- S. Y. Park, I. A. Jang, M. A. Lee, Y. J. Kim, S. H. Chun, and M. H. Park. (2016). Screening for chro- mosomal abnormalities using combined test in the first trimester of pregnancy, Obstetrics & Gynecology Science, vol. 59, no. 5, p. 357.
- D.W.Bianchi, R.L.Parker, J.Wentworth, R.Madankumar, C.Saffer, A.F.D, J.A.Craig, D.I. Chudova, P. L. Devers, K. W. Jones, K. Oliver, R. P. Rava, A. J. Sehnert. (2014). DNA Sequencing versus Standard Prenatal Aneuploidy Screening, New England Journal of Medicine, vol. 370, pp. 799–808.
- C. G. Artieri, C. Haverty, E. A. Evans, J. D. Goldberg, I. S. Haque, Y. Yaron, and D. Muzzey. (2017). Noninvasive prenatal screening at low fetal fraction: comparing whole-genome sequencing and single-nucleotide polymorphism methods, Prenatal Diagnosis, vol. 37, pp. 482–490.
- P. Benn, E. Valenti, S. Shan, K. Martin, Z. Demko. (2018). Factors Associated With Informative Redraw After an Initial No Result in Noninvasive Prenatal Testing, Obstetrics & Gynecology, vol. 132, pp. 428-435.
Citation: Malena Götte., et al. (2020). Performances of a Snp-Based Noninvasive Prenatal Test. Journal of Gynaecology and Paediatric Care 2(2). DOI: 10.5281/zenodo.3953715
Copyright: © 2020 Malena Götte. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.