Research Article
Volume 2 Issue 2 - 2020
Molecular Characterization of Cultivated Sugarcane Varieties on Tabasco, Mexico.
1Colegio de Postgraduados, Campus Tabasco. Periférico Carlos A. Molina s/n, H. Cárdenas 86500, Tabasco, México
2Colegio de Postgraduados, Campus Montecillo. Carretera México-Texcoco km 36.5 Montecillo 56230. Estado de México
3Estudiante de del Program de Doctorado en Ciencias Agrícolas en el Tropico del Colegio de Postgraduados, Campus Tabasco. Periférico Carlos A. Molina s/n, H. Cárdenas 86500, Tabasco, México
2Colegio de Postgraduados, Campus Montecillo. Carretera México-Texcoco km 36.5 Montecillo 56230. Estado de México
3Estudiante de del Program de Doctorado en Ciencias Agrícolas en el Tropico del Colegio de Postgraduados, Campus Tabasco. Periférico Carlos A. Molina s/n, H. Cárdenas 86500, Tabasco, México
*Corresponding Author: González-Jiménez V, Colegio de Postgraduados, Campus Tabasco. Periférico Carlos A. Molina s/n, H. Cárdenas 86500, Tabasco, México.
Received: May 04, 2020; Published: May 14, 2020
Abstract
In recent years different types of morphological, biochemical and molecular methods have been used to characterize plant varieties. The aim of this study was to characterize molecular genetic variability in 12 cultivated sugarcane (Saccharum spp.) varieties in the state of Tabasco Mexico by AFLP. Young leaves of plants growing in the field were selected for experiment. After testing 12 combinations of primers, the combination E-ACC/M-CTA produced from 72 to 1353 bp polymorphic fragments. The dendrogram revealed two distinct groups of varieties and a variety different from both groups. The varieties C 87-51, ATM 96-40, B 4362, Mex 69-290, Mex 57-1285 and Mex 91-130 integrated the Group I. Such varieties formed a cluster with a similarity of 0.77% among them. The varieties RD 75-11, Mex 79-431, SP 70-1284, Mex 59-32 and CP 72-2086 integrated the Group II. Such varieties formed another cluster with a similarity of 0.70% among them. The variety Mex 68-P-23 was different from the other 11 varieties.
Key words: DNA; AFLP; Molecular markers
Introduction
The increase in world sugar production is due to the introduction of improved cultivars (Jackson, 2005). The modern cultivars of sugar cane (Saccharum spp) currently planted in the world resulted from the first interspecific cross among S. officinarum, S. spontaneum and S. barberi carried out at the beginning of the last century (Grivet and Arruda, 2002). Several authors as Baksha et al. (2002) and Lu et al. (1994) have pointed out the need to create new sugarcane germplasm with a higher sugar yield, high resistance to abiotic and to biotic factors and with easy agronomic management.
Saccharum officinarum is an octoploid species with a chromosomal number 2n = 70 - 140 (Irvine, 1999). Therefore, and due to phenomena of euploidy and aneuploidy, commercial clones could be the product of numerous hybridizations with high chromosome numbers 2n = 100 - 130 (Grivet and Arruda, 2002). Thus, in the generation of new sugarcane varieties the characterization of varieties is an important aspect.
Recently, progress has been made in searching for genes of interest to characterize plant varieties by using various types of markers. The biochemists and DNA-based markers are the most widely used. Some of them are: Restriction Fragment Length Polymorphism (RFLP), Random Amplification of Polymorphic DNA (RAPD), Amplified Fragment Length Polymorphism (AFLP) and Microsatellites or Simple Repeated Sequences (SSR). These markers allow the identification and isolation of genes of interest through DNA amplification by the Polymerase Chain Reaction (PCR) technique. They are currently being used in the genetic tracing of sugarcane (Hoarau et al., 2001; Rossi et al., 2003), as well as in deducing inferences about genetic variability and interrelationships between genotypes at the level of DNA (Lima et al., 2002; Cordeiro et al., 2003), among which the construction of genetic maps in commercial varieties stands out (Aitken et al., 2005).
The AFLP marker combines RFLP and PCR technique and consists of the amplification of multiple arbitrary regions of the genome (Vos et al., 1995). The use of AFLP has been successful to get distributed molecular markers in eukaryotic genomes and in cultivated plants that have a low rate of DNA polymorphism, as well as in the detection and evaluation of genetic variation in germplasm collections and biodiversity studies (Vuylsteke et al., 2000). Therefore, AFLP marker was chosen to carry out the molecular characterization in varieties of sugar cane from the state of Tabasco.
Materials and Methods
Sugarcane varieties
Twelve sugarcane varieties from the germplasm bank located in the experimental field of the Postgraduate College - Campus Tabasco were used (Table 1).
Twelve sugarcane varieties from the germplasm bank located in the experimental field of the Postgraduate College - Campus Tabasco were used (Table 1).
| Variety | Parents | Country of origin |
| C 87-51 | Co 281 x POJ 2878 | Cuba |
| Mex 57-1285 | CP 52-43 x CB 45-6 | Mexico |
| Mex 59-32 | B 35-187 x CP 34-120 | Mexico |
| Mex 91-130 | Mex 57-280 x Mex 72-161 | Mexico |
| ATM 96-40 | SP 70-6180 x Mex 79-431 | Mexico |
| RD 75-11 | CB 38-22 x CP 57-603 | República Dominicana |
| Mex 79-431 | Co 421 x Mex 57-473 | Mexico |
| B 4362 | B 37-161 x POJ 2878 | Barbados |
| Mex 69-290 | Mex 56-476 x Mex 53-142 | Mexico |
| SP 70-1284 | CB 41-76 x | Brasil |
| CP 72-2086 | CP 62-374 x CP 63-588 | Estados Unidos |
| Mex 68-P-23 | Mex 59-84 x | Mexico |
Table 1: Cultivated sugarcane varieties (Saccharum spp.) in the state of Tabasco Mexico, used to determine its genetic variability.
Sugarcane plants growing on field were used to sample young leaves. Samples were collected according to the protocol of the applied molecular genetics laboratory of the International Maize and Wheat Improvement Center (CIMMYT, 2006).
DNA extraction
Baindrige et al. (1990). DNA concentration for each sample was determined by the spectrometric method. Then the concentrations of all samples were equalized to 100 ng µl-1. The AFLPs procedure was performed following the recommendations indicated in the AFLP®Analysis System I Kit and AFLP® Starter Primer Kit (InvitrogenTM, Carlsbad, CA).
Baindrige et al. (1990). DNA concentration for each sample was determined by the spectrometric method. Then the concentrations of all samples were equalized to 100 ng µl-1. The AFLPs procedure was performed following the recommendations indicated in the AFLP®Analysis System I Kit and AFLP® Starter Primer Kit (InvitrogenTM, Carlsbad, CA).
Genomic DNA digestion
Genomic DNA was digested using restriction enzymes EcoRI and MseI. A mixture of 5X Buffer reaction (5 µl), tomato control DNA (100 ng in 5 µl (2.5 µl)), DNA sample (250 ng in 18 μl), EcoRI/MseI (2 μl), distilled water (15.5 μl) for a final volume of 25 μl. It was capped at 37°C during 2h.
Genomic DNA was digested using restriction enzymes EcoRI and MseI. A mixture of 5X Buffer reaction (5 µl), tomato control DNA (100 ng in 5 µl (2.5 µl)), DNA sample (250 ng in 18 μl), EcoRI/MseI (2 μl), distilled water (15.5 μl) for a final volume of 25 μl. It was capped at 37°C during 2h.
Ligation of adapters
For ligation, 24 µl of ligation solution adapters and 1 µl of T4 DNA ligase were used to prepare a 1:5 dilution.
For ligation, 24 µl of ligation solution adapters and 1 µl of T4 DNA ligase were used to prepare a 1:5 dilution.
Pre-amplification
The pre-amplification was performed with a 1:10 dilution of the Primer Mix (40 µl), 10X PCR buffer plus Mg (5.0 µl), Taq DNA polymerase (1 µl). The resulting solution was taken to a DNA thermocycler (Bio-Rad Engine Peltier Thermal Cycler), previously programmed for 20 cycles at 94°C during 30s, 56°C during 1 min and 72°C during 1 min. 1:10 dilutions with water were prepared for AFLP (Sigma) from the pre- amplification.
The pre-amplification was performed with a 1:10 dilution of the Primer Mix (40 µl), 10X PCR buffer plus Mg (5.0 µl), Taq DNA polymerase (1 µl). The resulting solution was taken to a DNA thermocycler (Bio-Rad Engine Peltier Thermal Cycler), previously programmed for 20 cycles at 94°C during 30s, 56°C during 1 min and 72°C during 1 min. 1:10 dilutions with water were prepared for AFLP (Sigma) from the pre- amplification.
Selective amplification
According to the manufacturer's recommendations, possible combinations of primers were sought to find the ones that provide the best banding pattern and thus be able to differentiate the 12 varieties of sugarcane (Table 2).
According to the manufacturer's recommendations, possible combinations of primers were sought to find the ones that provide the best banding pattern and thus be able to differentiate the 12 varieties of sugarcane (Table 2).
Data analysis
The bands, from the electrophoretic patterns obtained from the molecular tests, were evaluated in a binary way (0 absence, 1 presence). The coefficient of genetic similarity between each pair of genotype was calculated from the matrices of the original data by using only the polymorphic bands. The NTSys PC 2.0 statistical package (dendrogram and genetic distance matrix) was used.
The bands, from the electrophoretic patterns obtained from the molecular tests, were evaluated in a binary way (0 absence, 1 presence). The coefficient of genetic similarity between each pair of genotype was calculated from the matrices of the original data by using only the polymorphic bands. The NTSys PC 2.0 statistical package (dendrogram and genetic distance matrix) was used.
| Combination Number | Primer combination Eco RI / Mse I |
| 1 | E-AAC/M-CAC |
| 2 | E-AAG/M-CTC |
| 3 | E-ACA/M-CTT |
| 4 | E-ACC/M-CTA |
| 5 | E-AAC/M-CAG |
| 6 | E-AAC/M-CTG |
| 7 | E-ACC/M-CTG |
| 8 | E-ACG/M-CAC |
| 9 | E-ACG/M-CAA |
| 10 | E-ACT/M-CTC |
| 11 | E-AGC/M-CAT |
| 12 | E-AGG/M-CAT |
Table 2: Primer combinations used for the molecular characterization of 12 cultivated sugarcane varieties (Saccharum spp.) in the state of Tabasco, Mexico.
Results and Discussion
The amplification products evaluated to visualize and carry out analysis of polymorphisms with AFLP markers allow us to observe genetic differences among the analyzed varieties. After performing 12 combinations of these amplifiers, the E-ACC/M-CTA combination showed the highest polymorphism among varieties. The size of the polymorphic fragments detected with this primer combination ranged from 72 to 1353 bp (Figure 1).
The low polymorphism detected indicates the narrow genetic base of the 12 evaluated varieties cultivated in the Tabasco state Mexico. This narrow genetic base may be due to the high polyploidy and the frequent aneuploidy of the genus Saccharum (Grivet and Arruda, 2002). Moreorver sugarcane genome is complex ranging from 2500-4000 million base pairs (Mbp/1C) (Arumuganathan and Earle, 1991). Only the Saccharum offcinarum chloroplast DNA genome is 141182 bp (Asano et al., 2004; Calsa et al., 2004).
Another reason for the low polymorphism observed may be that the crosses between genotypes focus mainly on the generation of varieties with resistance to pests and diseases (Harvey et al., 1994). These crosses have been successful and have originated varieties of sugarcane more productive but with a marked reduction in its genetic base (Jannoo et al., 1999).
The AFLP technique has been successful in sugarcane in several countries. In Brazil, Lima et al. (2002) detected an average of 50% polymorphism among 79 cultivars. In India, Selvi et al. (2006) reported an average of 52% polymorphism among 28 cultivars by using 12 primer combinations.
In Mexico, Rodríguez et al. (2005) characterized the 15 most sowed sugarcane varieties by using 12 AFLP combinations. 55.3% of their 884 generated markers were polymorphic, the Mex 73-523 y Mex 68-P-23 varieties were in the dendrogram further branches and were apart to the other varieties.
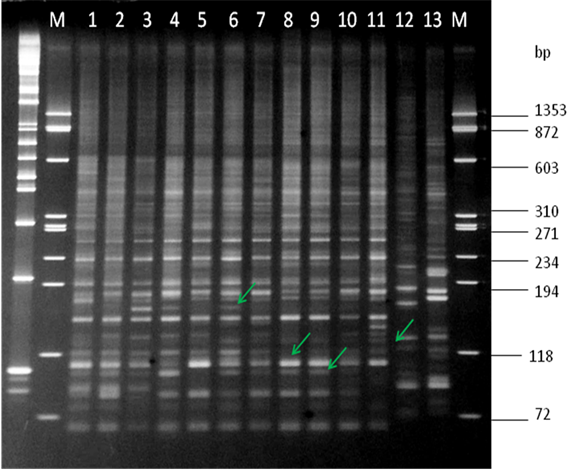
Figure 1: E-ACC/M-CTA combination. Varieties: 1) C 87-51; 2) Mex 57-1285; 3) Mex 59-32; 4) Mex 91-130; 5) ATM 96-40; 6) RD 75-11; 7) Mex 79-431; 8) B 4362; 9) Mex 69-290; 10) SP 70-1284; 11) CP 72-2086; 12) Mex 68-P-23; 13) control +; M) Φ X174 / HáeIII marker. (Arrows indicate dispersion of polymorphic bands).
Raboin et al. (2008) studied 72 Saccharum spp clones using the AFLP technique. They found 1537 polymorphic markers with 42 combinations. Among these markers, only 463 were located on the genetic map.
The dendrogram revealed three different groups of Saccharum spp.and a variety different from both groups (Figure 2). The varieties C 87-51, ATM 96-40, B 4362, Mex 69-290, Mex 57-1285 and Mex 91-130 integrated the Group I. Such varieties presented a 0.77% genetic similarity, i.e. these materials showed more genetic closeness among them compared to the varieties of group II. Group II included the varieties RD 75-1, Mex 79-431, SP 70-1284, Mex 59-32 and CP 72-2086. These varieties formed a conglomerate with 70% genetic similarity among them.
The Mex 68-P-23 variety was the one with the least genetic similarity (22%) compared to the rest of the analyzed varieties.
Rodríguez et al. (2005) reported that the 15 sugarcane varieties most used in the production in Mexico formed a narrow genetic population and that the Mex 68-P-23 variety had a greater genetic distance. The latter could indicate an increase in genetic variability, which is essential for genetic improvement. These and our results agree to the multiple studies citing out the narrow genetic base of modern cultivars (Grivet et al., 1996; Canales et al., 2003).
The sugarcane genotypes study has shown that there is a small degree of DNA diversity among modern varieties (Arro, 2005). In Cuba, Arencibia et al. (2006) found that the current commercial varieties of sugarcane present low genetic variability among them.
The above and our results can be explained by the “nobilization process”. This process mean that the crosses of the modern cultivars used as parents of the first crosses were backcrossed several times with S. officinarum. As result of “nobilization” the new hybrids showed a reduction in their genetic base (Deren, 1995; Jannoo et al., 1999).
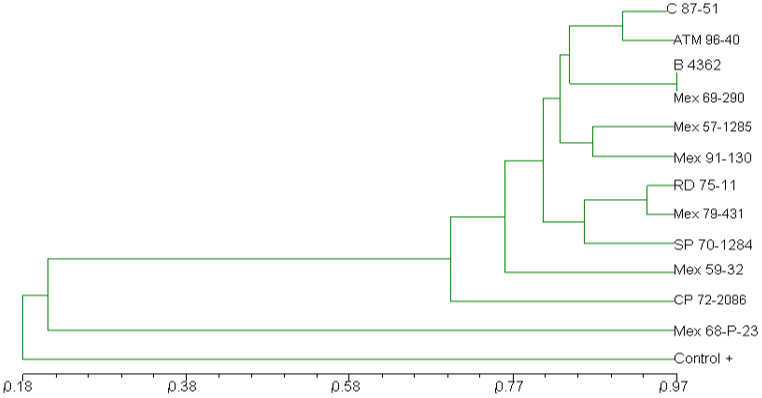
Figure 2: Cluster analysis based on the genetic similarity AFLP of 12 cultivated sugarcane varieties in the state of Tabasco Mexico by the NTSYS PC program.
Conclusions
The results confirm the usefulness of AFLP to obtain the necessary number of markers for genome analysis, as well as to characterize and to detect polymorphisms of 12 sugarcane varieties in the state of Tabasco.
The E-ACC / M-CTA combination was the one that produced the largest polymorphic fragments. A small degree of diversity at the DNA level among the cultivated sugarcane varieties in the state of Tabasco Mexico was found. The Mex 68-P-23 variety presented a faraway genetic distance with respect to the rest of the analyzed varieties.
Acknowledgments
Thanks are given to the Postgraduate College - Campus Tabasco and to the Priority Research Line 5 (LPI-5) Microbial, plant and animal biotechnology for the financial support to carry out this work. The first author thanks to the National Council of Science and Technology for the study grant of the Master of Science.
Thanks are given to the Postgraduate College - Campus Tabasco and to the Priority Research Line 5 (LPI-5) Microbial, plant and animal biotechnology for the financial support to carry out this work. The first author thanks to the National Council of Science and Technology for the study grant of the Master of Science.
References
- Aitken KA, Jackson PA, McIntyre CL. (2005). A combination of AFLP and SSR markers provide extensive map coverage and identification of homo (eo) logouts linkage groups in a sugarcane cultivar. Theor Appl Genet. 110: 789-701.
- Arencibia A, Delgado M, Jorge H, Coto O, Ibis J, García H. (2006). Molecular characterization of cuban sugarcane varieties (Saccharum spp) by AFLP. Revista Fitotecnia Mexicana. 29(1): 19-25
- Arro JA. (2005). Genetic diversity among sugarcane clones using Target Region Amplification Polymorphism (TRAP) markers and pedigree relationships. Masters Thesis submitted to Louisiana State University, Baton Rouge. 78 p
- Arumuganathan K, Earle ED. (1991). Nuclear DNA content of some important plant species. Plant Molecular Biology Reporter 9: 208-218.
- Asano T, Tsudzuki T, Takahashi S, Shimada H, Kadowaki K. (2004). Complete nucleotide sequence of the sugarcane (Saccharum officinarum) chloroplast genome: a comparative analysis of four monocot chloroplast genomes.DNAResearch 11(2): 93-99.
- Baindrige BW, Spreadbury CL, Scalise FG, Cohen J. (1990). Improved methods for the preparation of high molecular weight DNA from large and small scale cultures of filamentous fungi. Microbiology Letter 66: 113-118.
- Baksha R, Alam R, Karim MZ, Paul SK, Hossain, MA, Miah MAS, Rahman ABMM. (2002). In vitro shoot tip culture of sugarcane (Saccharum officinarum) Variety Isd 28. Biotechnology. 1(2-4): 67-72
- Calsa JT, Carraro DM, Benatti MR, Barbosa AC, Kitajima JP, Carrer H. (2004). Structural features and transcript-editing analysis of sugarcane (Saccharum officinarum L.) chloroplast genome. Curr Genet. 46: 366-373.
- Canales E, Coto O, Cornide MT. (2003). Variación genética e identificación de cultivares cubanos de caña de azúcar mediante RFLP. Revista CENIC Ciencias Biológicas. 34(3): 129-136
- CIMMYT (Centro Internacional de Mejoramiento de Maíz y Trigo). (2006). Protocolos de laboratorio: Laboratorio de Genética Molecular Aplicada del CIMMYT. 3ra. ed. México. D. F. p. 92
- Cordeiro GM, Pan YB, and Henry RJ. (2003). Sugarcane microsatellites for the assessment of genetic diversity in sugarcane germplasm. Plant Science 165: 181-189.
- Deren CW. (1995). Genetic Base of U.S. Mainland Sugarcane. Crop Science 35: 1195-1199.
- Grivet L, Arruda P. (2002). Sugarcane genomics: depicting the complex genome of an important tropical crop. Curr Opin Plant Biol5(2): 122-127.
- Grivet L, D’Hont A, Rodrigues D, Feldmann P, Lanaud C, Glaszmann JC. (1996). RFLP mapping in cultivated sugarcane (Saccharum spp.): genome organization in a highly polyploid and aneuploid interspecific hybrid. Genetics 142: 987-1000.
- Harvey H, Huckett BI, Botha FC. (1994). Use of Polymerase Chain Reaction and Random Amplification of Polymorphic DNAs for the determination of genetic distances between 21 sugarcane varieties. Proceedings of South African Sugar Association. 68: 36-40.
- Hoarau JY, Offmann B, D’Hont A, Risterucci AM, Roques D, Glaszmann JC, Grivet L. (2001). Genetic dissection of a modern sugarcane cultivar (Saccharum spp.) I. Genome mapping with AFLP markers. Theor Appl Genet103: 84–97.
- Irvine JE. (1999). Saccharum species as horticultural classes. Theor Appl Genet. 98: 186-194.
- Jackson PA. (2005). Breeding for improved sugar content in sugarcane. Field Crops Research 92(2-3): 277-290.
- Jannoo NL, Grivet M, Seguin F, Paulet R, Domaingue PS, Rao A, Dookun A, D'Hont A, Glaszmann JC. (1999). Molecular investigation of the genetic base of sugarcane cultivars. Theor Appl Genet 99: 171-184.
- Lima MLA, Garcia AAF, Oliveira KM, Matsuoka SH, Arizono CL, De Souza Jr AP, De Souza AP. (2002). Analysis of genetic similarity detected by AFLP and coefficient of parentage among genotypes of sugarcane (Saccharum spp.) Theor Appl Genet 104: 30-38.
- Lu YH, D'Hont A, Walker DJT, Rao PS, Feldmann P, Glaszmann JC. (1994). Relationships among ancestral species of sugarcane revealed with RFLP using single-copy maize nuclear probes. Euphytica 78: 7-18.
- Raboin LM, Pauquet J, Butterfield M, D’Hont A, Glaszmann JC. (2008). Analysis of genome-wide linkage disequilibrium in the highly polyploid sugarcane. Theor Appl Genet 116: 701-714.
- Rodríguez HAM, Castillo CMA, Flores BEP. (2005). Genetic Diversity of the most important sugarcane cultivars in México. e–Gnosis 3: 1-10.
- Rossi M, Araujo PG, Paulet F, Garsmeur O, Dias VM, Chen H, Van- Sluys MA, D’Hont AD. (2003). Genomic distribution and characterization of EST-derived resistance gene analogs (RGAs) in sugarcane. Molecular Genetics and Genomics. 269: 406-419.
- Selvi A, Nair NV, Noyer JL, Singh NK, Balasundaram N, Bansal KC, Koundal KR, Mohapatra T. (2006). AFLP analysis of the phenetic organization and genetic diversity in the sugarcane complex, Saccharum and Erianthus. Genetic Resources and Crop Evolution 53(4): 831-842.
- Vos P, Hogers R, Bleeker M, Reijans MV, Der LT, Hornes M. (1995). AFLP a new concept for DNA fingerprinting. Nucleic Acids Research 23: 4407-4414.
- Vuylsteke M, Mank R, Brugmans B, Stam P, Kuiper M. (2000). Further characterization of AFLP data as a tool in genetic diversity assessments among maize (Zea mays L.) inbred lines. Mol. Breeding. 6(3): 265-276.
Citation: Gonzalez-Jimenez V. et al. (2020). Molecular Characterization of Cultivated Sugarcane Varieties on Tabasco, Mexico. Journal of Biotechnology and Immunology 2(2).
Copyright: © 2020 Gonzalez-Jimenez V. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
