Research Article
Volume 1 Issue 1 - 2019
Methods of study in Molecular Agrobacterium and genes
Assistant Professor in Semnan University –Iran
*Corresponding Author: Hamid Kheyrodin, Assistant Professor in Semnan University –Iran.
Received: June 07, 2019; Published: July 08, 2019
Abstract
Genetic transformation of plant cells by Agrobacterium tumefaciens is the only known natural example of trans-kingdom DNA transfer. In nature, Agrobacterium introduces several oncogenic genes into the host plant, leading to formation of tumors [1], and in the laboratory this microorganism is used widely for plant genetic engineering [2, 3]. Agrobacterium infection requires the presence of two genetic components located on the bacterial tumor-inducing (Ti) plasmid: the transferred DNA (T-DNA), which is introduced into the plant cell genome, and the virulence (vir) region composed of seven loci—virA, virB, virC, virD, virE, virG, and virH encoding most components of the protein apparatus for T-DNA transfer. In addition, several bacterial chromosomal virulence (chv) genes participate in the early stages of Agrobacterium attachment to the plant cells [reviewed in refs. 4 and 5–7].
We suggested that Agrobacterium tumefaciens is a soil phytopathogen that elicits neoplastic growths on the host plant species. In nature, however, Agrobacterium also may encounter organisms belonging to other kingdoms such as insects and animals that feed on the infected plants.
Keywords: Agrobacterium; Cell; Plant; Genes
Introduction
Conjucation
To be virulent, the bacterium must contain a tumour-inducing plasmid (Ti plasmid or pTi), of 200 kbp, which contains the T-DNA and all the genes necessary to transfer it to the plant cell. Many strains of A. tumefaciens do not contain a pTi. Since the Ti plasmid is essential to cause disease, prepenetration events in the rhizosphere occur to promote bacterial conjugation - exchange of plasmids amongst bacteria. In the presence of opines, A. tumefaciens produces a diffusible conjugation signal called 30C8HSL or the Agrobacterium autoinducer. This activates the transcription factor TraR, positively regulating the transcription of genes required for conjugation.
To be virulent, the bacterium must contain a tumour-inducing plasmid (Ti plasmid or pTi), of 200 kbp, which contains the T-DNA and all the genes necessary to transfer it to the plant cell. Many strains of A. tumefaciens do not contain a pTi. Since the Ti plasmid is essential to cause disease, prepenetration events in the rhizosphere occur to promote bacterial conjugation - exchange of plasmids amongst bacteria. In the presence of opines, A. tumefaciens produces a diffusible conjugation signal called 30C8HSL or the Agrobacterium autoinducer. This activates the transcription factor TraR, positively regulating the transcription of genes required for conjugation.
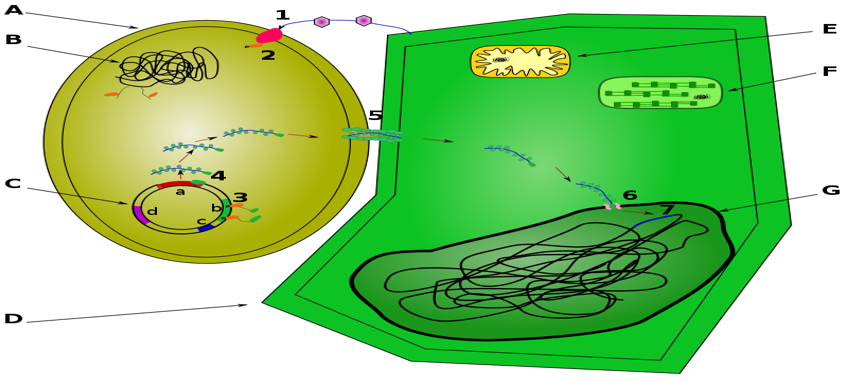
Figure 1: A: Agrobacterium tumefaciens B: Agrobacterium genome C: Ti Plasmid: a: T-DNA , b: Vir genes , c: Replication origin , d: Opines catabolism genes D: Plant cell E: Mitochondria F: Chloroplast G: Nucleus
Method of Infection
A. tumefaciens infects the plant through its Ti plasmid. The Ti plasmid integrates a segment of its DNA, known as T-DNA, into the chromosomal DNA of its host plant cells. A. tumefaciens has flagella that allow it to swim through the soil towards photoassimilates that accumulate in the rhizosphere around roots. Some strains may chemotactically move towards chemical exudates from plants, such as acetosyringone and sugars. The former is recognised by the VirA protein, a transmembrane protein encoded in the virA gene on the Ti plasmid. Sugars are recognised by the chvE protein, a chromosomal gene-encoded protein located in the periplasmic space. [8]
A. tumefaciens infects the plant through its Ti plasmid. The Ti plasmid integrates a segment of its DNA, known as T-DNA, into the chromosomal DNA of its host plant cells. A. tumefaciens has flagella that allow it to swim through the soil towards photoassimilates that accumulate in the rhizosphere around roots. Some strains may chemotactically move towards chemical exudates from plants, such as acetosyringone and sugars. The former is recognised by the VirA protein, a transmembrane protein encoded in the virA gene on the Ti plasmid. Sugars are recognised by the chvE protein, a chromosomal gene-encoded protein located in the periplasmic space. [8]
At least 25 vir genes on the Ti plasmid are necessary for tumor induction. In addition to their perception role, virA and chvE induce other vir genes. The virA protein has auto kinase activity: it phosphorylates itself on a histidine residue. Then the virA protein phosphorylates the virG protein on its aspartate residue. The virG protein is a cytoplasmic protein produced from the virG Ti plasmid gene. It is a transcription factor, inducing the transcription of the vir operons. The chvE protein regulates the second mechanism of the vir genes' activation. It increases VirA protein sensitivity to phenolic compounds. [8] Attachment is a two-step process. Following an initial weak and reversible attachment, the bacteria synthesize cellulose fibrils that anchor them to the wounded plant cell to which they were attracted. Four main genes are involved in this process: chvA, chvB, pscA, and att. The products of the first three genes apparently are involved in the actual synthesis of the cellulose fibrils. These fibrils also anchor the bacteria to each other, helping to form a microcolony.
VirC, the most important virulent gene, is a necessary step in the recombination of illegitimate recolonization. It selects the section of the DNA in the host plant that will be replaced and it cuts into this strand of DNA.
After production of cellulose fibrils, a calcium-dependent outer membrane protein called rhicadhesin is produced, which also aids in sticking the bacteria to the cell wall. Homologues of this protein can be found in other rhizobia. Currently, there are several reports on standardisation of protocol for the Agrobacterium-mediated transformation. Effect of different parameters like, infection time, acetosyringone, DTT, cysteine have been studied in soybean (Glycine max) [9]
Possible plant compounds that initiate Agrobacterium to infect plant cells: [10]
- Acetosyringone and other phenolic compounds
- alpha-Hydroxyacetosyringone
- Catechol
- Ferulic acid
- Gallic acid
- p-Hydroxybenzoic acid
- Protocatechuic acid
- Pyrogallic acid
- Resorcylic acid
- Sinapinic acid
- Syringic acid
- Vanillin
Genes in the T-DNA
Hormones
To cause gall formation, the T-DNA encodes genes for the production of auxin or indole-3-acetic acid via the IAM pathway. This biosynthetic pathway is not used in many plants for the production of auxin, so it means the plant has no molecular means of regulating it and auxin will be produced constitutively. Genes for the production of cytokinins are also expressed. This stimulates cell proliferation and gall formation.
Hormones
To cause gall formation, the T-DNA encodes genes for the production of auxin or indole-3-acetic acid via the IAM pathway. This biosynthetic pathway is not used in many plants for the production of auxin, so it means the plant has no molecular means of regulating it and auxin will be produced constitutively. Genes for the production of cytokinins are also expressed. This stimulates cell proliferation and gall formation.
Opines
The T-DNA contains genes for encoding enzymes that cause the plant to create specialized amino acid derivatives which the bacteria can metabolize, called opines. [11] Opines are a class of chemicals that serve as a source of nitrogen for A. tumefaciens, but not for most other organisms. The specific type of opine produced by A. tumefaciens C58 infected plants is nopaline (Escobar et al., 2003). Two nopaline type Ti plasmids, pTi-SAKURA and pTiC58, were fully sequenced. A. tumefaciens C58, the first fully sequenced pathovar, was first isolated from a cherry tree crown gall. The genome was simultaneously sequenced by Goodner et al. [12] and Wood et al. [13] in 2001. The genome of A. tumefaciens C58 consists of a circular chromosome, two plasmids, and a linear chromosome. The presence of a covalently bonded circular chromosome is common to Bacteria, with few exceptions. However, the presence of both a single circular chromosome and single linear chromosome is unique to a group in this genus. The two plasmids are pTiC58, responsible for the processes involved in virulence, and pAtC58, dubbed the "cryptic" plasmid. [12,13] The pAtC58 plasmid has been shown to be involved in the metabolism of opines and to conjugate with other bacteria in the absence of the pTiC58 plasmid. [14] If the pTi plasmid is removed, the tumor growth that is the means of classifying this species of bacteria does not occur.
The T-DNA contains genes for encoding enzymes that cause the plant to create specialized amino acid derivatives which the bacteria can metabolize, called opines. [11] Opines are a class of chemicals that serve as a source of nitrogen for A. tumefaciens, but not for most other organisms. The specific type of opine produced by A. tumefaciens C58 infected plants is nopaline (Escobar et al., 2003). Two nopaline type Ti plasmids, pTi-SAKURA and pTiC58, were fully sequenced. A. tumefaciens C58, the first fully sequenced pathovar, was first isolated from a cherry tree crown gall. The genome was simultaneously sequenced by Goodner et al. [12] and Wood et al. [13] in 2001. The genome of A. tumefaciens C58 consists of a circular chromosome, two plasmids, and a linear chromosome. The presence of a covalently bonded circular chromosome is common to Bacteria, with few exceptions. However, the presence of both a single circular chromosome and single linear chromosome is unique to a group in this genus. The two plasmids are pTiC58, responsible for the processes involved in virulence, and pAtC58, dubbed the "cryptic" plasmid. [12,13] The pAtC58 plasmid has been shown to be involved in the metabolism of opines and to conjugate with other bacteria in the absence of the pTiC58 plasmid. [14] If the pTi plasmid is removed, the tumor growth that is the means of classifying this species of bacteria does not occur.
Biotechnological uses
The DNA transmission capabilities of Agrobacterium have been vastly explored in biotechnology as a means of inserting foreign genes into plants. Marc Van Montagu and Jeff Schell, (University of Ghent and Plant Genetic Systems, Belgium) discovered the gene transfer mechanism between Agrobacterium and plants, which resulted in the development of methods to alter the bacterium into an efficient delivery system for genetic engineering in plants. [15] The plasmid T-DNA that is transferred to the plant is an ideal vehicle for genetic engineering. [16] This is done by cloning a desired gene sequence into the T-DNA that will be inserted into the host DNA. This process has been performed using firefly luciferase gene to produce glowing plants. This luminescence has been a useful device in the study of plant chloroplast function and as a reporter gene. [17] It is also possible to transform Arabidopsis thaliana by dipping flowers into a broth of Agrobacterium: the seed produced will be transgenic. Under laboratory conditions, the T-DNA has also been transferred to human cells, demonstrating the diversity of insertion application. [18]
The DNA transmission capabilities of Agrobacterium have been vastly explored in biotechnology as a means of inserting foreign genes into plants. Marc Van Montagu and Jeff Schell, (University of Ghent and Plant Genetic Systems, Belgium) discovered the gene transfer mechanism between Agrobacterium and plants, which resulted in the development of methods to alter the bacterium into an efficient delivery system for genetic engineering in plants. [15] The plasmid T-DNA that is transferred to the plant is an ideal vehicle for genetic engineering. [16] This is done by cloning a desired gene sequence into the T-DNA that will be inserted into the host DNA. This process has been performed using firefly luciferase gene to produce glowing plants. This luminescence has been a useful device in the study of plant chloroplast function and as a reporter gene. [17] It is also possible to transform Arabidopsis thaliana by dipping flowers into a broth of Agrobacterium: the seed produced will be transgenic. Under laboratory conditions, the T-DNA has also been transferred to human cells, demonstrating the diversity of insertion application. [18]
The mechanism by which Agrobacterium inserts materials into the host cell is by a type IV secretion system which is very similar to mechanisms used by pathogens to insert materials (usually proteins) into human cells by type III secretion. It also employs a type of signaling conserved in many Gram-negative bacteria called quorum sensing. This makes Agrobacterium an important topic of medical research, as well.
Natural genetic transformation
Natural genetic transformation in bacteria is a sexual process involving the transfer of DNA from one cell to another through the intervening medium, and the integration of the donor sequence into the recipient genome by homologous recombination. A. tumefaciens can undergo natural transformation in soil without any specific physical or chemical treatment. [19]
Natural genetic transformation in bacteria is a sexual process involving the transfer of DNA from one cell to another through the intervening medium, and the integration of the donor sequence into the recipient genome by homologous recombination. A. tumefaciens can undergo natural transformation in soil without any specific physical or chemical treatment. [19]
Disease Cycle
Agrobacterium tumefaciens overwinters in infested soils. Agrobacterium species live predominantly saprophytic lifestyles, so it's common even for plant parasitic species of this genus to survive in the soil for lengthy periods of time, even without host plant presence. [20] When there is a host plant present, however, the bacteria enter the plant tissue via recent wounds or natural openings of roots or stems near the ground. These wounds may be caused by cultural practices, grafting, insects, etc. Once the bacteria have entered the plant, they occur intercellularly and stimulate surrounding tissue to proliferate due to cell transformation. Agrobacterium performs this control by inserting the plasmid T-DNA into the plant's genome. See above for more details about the process of plasmid DNA insertion into the host genome. Excess growth of the plant tissue leads to gall formation on the stem and roots. These tumors exert significant pressure on the surrounding plant tissue, which causes this tissue to become crushed and/or distorted. The crushed vessels lead to reduced water flow in the xylem. Young tumors are soft and therefore vulnerable to secondary invasion by insects and saprophytic microorganisms. This secondary invasion causes breakdown of the peripheral cell layers as well as tumor discoloration due to decay. Breakdown of the soft tissue leads to release of the Agrobacterium tumefaciens into the soil allowing it to restart the disease process with a new host plant. [21]
Agrobacterium tumefaciens overwinters in infested soils. Agrobacterium species live predominantly saprophytic lifestyles, so it's common even for plant parasitic species of this genus to survive in the soil for lengthy periods of time, even without host plant presence. [20] When there is a host plant present, however, the bacteria enter the plant tissue via recent wounds or natural openings of roots or stems near the ground. These wounds may be caused by cultural practices, grafting, insects, etc. Once the bacteria have entered the plant, they occur intercellularly and stimulate surrounding tissue to proliferate due to cell transformation. Agrobacterium performs this control by inserting the plasmid T-DNA into the plant's genome. See above for more details about the process of plasmid DNA insertion into the host genome. Excess growth of the plant tissue leads to gall formation on the stem and roots. These tumors exert significant pressure on the surrounding plant tissue, which causes this tissue to become crushed and/or distorted. The crushed vessels lead to reduced water flow in the xylem. Young tumors are soft and therefore vulnerable to secondary invasion by insects and saprophytic microorganisms. This secondary invasion causes breakdown of the peripheral cell layers as well as tumor discoloration due to decay. Breakdown of the soft tissue leads to release of the Agrobacterium tumefaciens into the soil allowing it to restart the disease process with a new host plant. [21]
Disease Management
Crown gall disease caused by Agrobacterium tumefaciens can be controlled by using various different methods. The best way to control this disease is to take preventative measures, such as sterilizing pruning tools so as to avoid infecting new plants. Performing mandatory inspections of nursery stock and rejecting infected plants as well as not planting susceptible plants in infected fields are also valuable practices. Avoiding wounding the crowns/roots of the plants during cultivation is important for preventing disease. In horticultural techniques in which multiple plants are joined together to grow as one, such as budding and grafting [22] these techniques lead to plant wounds. Wounds are the primary location of bacterial entry into the host plant. Therefore, it is advisable to perform these techniques during times of the year when Agrobacteria are not active. Control of root-chewing insects is also helpful to reduce levels of infection, since these insects cause wounds (aka bacterial entryways) in the plant roots. [21] It is recommended that infected plant material be burned rather than placed in a compost pile due to the bacteria's ability to live in the soil for many years. [23] Biological control methods are also utilized in managing this disease. During the 1970s and 1980s, a common practice for treating germinated seeds, seedlings, and rootstock was to soak them in a suspension of K84. K84 is composed of A. radiobacter, which is a species related to A. tumefaciens but is not pathogenic. K84 produces a bacteriocin (agrocin 84) which is an antibiotic specific against related bacteria, including A. tumefaciens. This method, which was successful at controlling the disease on a commercial scale, had the risk of K84 transferring its resistance gene to the pathogenic Agrobacteria. Thus, in the 1990s, use of a genetically engineering strain of K84, known as K-1026, was created. This strain is just as successful in controlling crown gall as K84 without the caveat of resistance gene transfer. [24]
Crown gall disease caused by Agrobacterium tumefaciens can be controlled by using various different methods. The best way to control this disease is to take preventative measures, such as sterilizing pruning tools so as to avoid infecting new plants. Performing mandatory inspections of nursery stock and rejecting infected plants as well as not planting susceptible plants in infected fields are also valuable practices. Avoiding wounding the crowns/roots of the plants during cultivation is important for preventing disease. In horticultural techniques in which multiple plants are joined together to grow as one, such as budding and grafting [22] these techniques lead to plant wounds. Wounds are the primary location of bacterial entry into the host plant. Therefore, it is advisable to perform these techniques during times of the year when Agrobacteria are not active. Control of root-chewing insects is also helpful to reduce levels of infection, since these insects cause wounds (aka bacterial entryways) in the plant roots. [21] It is recommended that infected plant material be burned rather than placed in a compost pile due to the bacteria's ability to live in the soil for many years. [23] Biological control methods are also utilized in managing this disease. During the 1970s and 1980s, a common practice for treating germinated seeds, seedlings, and rootstock was to soak them in a suspension of K84. K84 is composed of A. radiobacter, which is a species related to A. tumefaciens but is not pathogenic. K84 produces a bacteriocin (agrocin 84) which is an antibiotic specific against related bacteria, including A. tumefaciens. This method, which was successful at controlling the disease on a commercial scale, had the risk of K84 transferring its resistance gene to the pathogenic Agrobacteria. Thus, in the 1990s, use of a genetically engineering strain of K84, known as K-1026, was created. This strain is just as successful in controlling crown gall as K84 without the caveat of resistance gene transfer. [24]
Environment
Host, environment, and pathogen are extremely important concepts in regards to plant pathology. Agrobacteria have the widest host range of any plant pathogen, [25] so the main factor to take into consideration in the case of crown gall is environment. There are various conditions and factors that make for a conducive environment for A. tumefaciens when infecting its various hosts. The bacterium can't penetrate the host plant without an entry point such as a wound. Factors leading to wounds in plants include cultural practices, grafting, freezing injury, growth cracks, soil insects, and other animals in the environment causing damage to the plant. Consequently, in exceptionally harsh winters, it's common to have an increased incidence of crown gall due to the weather-related damage. [26] Along with this, there are methods of mediating infection of the host plant. For example, nematodes can act as a vector to introduce Agrobacterium into plant roots. More specifically, the root parasitic nematodes damage the plant cell, creating a wound for the bacteria to enter through. [27] Finally, temperature is a factor when considering A. tumefaciens infection. The optimal temperature for crown gall formation due to this bacterium is 22 degrees Celsius because of the thermosensitivity of T-DNA transfer. Tumor formation is significantly reduced at higher temperature conditions. [28]
Host, environment, and pathogen are extremely important concepts in regards to plant pathology. Agrobacteria have the widest host range of any plant pathogen, [25] so the main factor to take into consideration in the case of crown gall is environment. There are various conditions and factors that make for a conducive environment for A. tumefaciens when infecting its various hosts. The bacterium can't penetrate the host plant without an entry point such as a wound. Factors leading to wounds in plants include cultural practices, grafting, freezing injury, growth cracks, soil insects, and other animals in the environment causing damage to the plant. Consequently, in exceptionally harsh winters, it's common to have an increased incidence of crown gall due to the weather-related damage. [26] Along with this, there are methods of mediating infection of the host plant. For example, nematodes can act as a vector to introduce Agrobacterium into plant roots. More specifically, the root parasitic nematodes damage the plant cell, creating a wound for the bacteria to enter through. [27] Finally, temperature is a factor when considering A. tumefaciens infection. The optimal temperature for crown gall formation due to this bacterium is 22 degrees Celsius because of the thermosensitivity of T-DNA transfer. Tumor formation is significantly reduced at higher temperature conditions. [28]
Conclusion
Agrobacterium tumefaciens (updated scientific name Rhizobium radiobacter, synonym Agrobacterium radiobacter) [2-4] is the causal agent of crown gall disease (the formation of tumours) in over 140 species of eudicots. It is a rod-shaped, Gram-negative soil bacterium. [1] Symptoms are caused by the insertion of a small segment of DNA (known as the T-DNA, for 'transfer DNA', not to be confused with tRNA that transfers amino acids during protein synthesis, confusingly also called transfer RNA), from a plasmid, into the plant cell, [5] which is incorporated at a semi-random location into the plant genome. A. tumefaciens is an alphaproteobacterium of the family Rhizobiaceae, which includes the nitrogen-fixing legume symbionts. Unlike the nitrogen-fixing symbionts, tumor-producing Agrobacterium species are pathogenic and do not benefit the plant. The wide variety of plants affected by Agrobacterium makes it of great concern to the agriculture industry. [6]
Economically, A. tumefaciens is a serious pathogen of walnuts, grape vines, stone fruits, nut trees, sugar beets, horse radish, and rhubarb. tumefaciens grows optimally at 28°C. The doubling time can range from 2.5–4h depending on the media, culture format and level of aeration. [7] At temperatures above 30 °C, A. tumefaciens begins to experience heat shock and is likely to result in errors in cell division. [7]
Footnotes
This paper was submitted directly (Track II) to you. Also this work was supported by a grant to Semnan University from the Biological and Biotechnological Sciences Research.
This paper was submitted directly (Track II) to you. Also this work was supported by a grant to Semnan University from the Biological and Biotechnological Sciences Research.
References
- Hooykaas P J J, Beijersbergen A. Annu Rev Phytopathol. (1994): 32:157–179.
- Lessl M, Lanka E. Cell. (1994): 77:321–324.
- Citovsky V, Zupan J, Warnick D, Zambryski P. Science. (1992): 256:1802–1805.
- Rossi L, Hohn B, Tinland B. Proc Natl Acad Sci USA. (1996): 93:126–130
- Regensburg-Tuink A J G, Hooykaas P. Nature (London) (1993): 363:69–70.
- Offringa R, de Groot M J A, Haagsman H J, Does M P, van den Elzen P J M, Hooykaas P J J. EMBO J. (1990): 9:3077–3084.
- Paszkowski J, Baur M, Bogucki A, Potrykus I. EMBO J. (1988): 7:4021–4026.
- Matsumoto S, Ito Y, Hosoi T, Takahashi Y, Machida Y. Mol Gen Genet. (1990): 224:309–316.
- Gheysen G, Villarroel R, Van Montagu M. Genes Dev. (1991): 5:287–297.
- Mayerhofer R, Koncz-Kalman Z, Nawrath C, Bakkeren G, Crameri A, Angelis K, Redei G P, Schell J, Hohn B, Koncz C. EMBO J. (1991): 10:697–704.
- Bundock P, den Dulk-Ras A, Beijersbergen A, Hooykaas P J J. EMBO J. (1995): 14:3206–3214.
- Piers K L, Heath J D, Liang X, Stephens K M, Nester E W. Proc Natl Acad Sci USA. (1996): 93:1613–1618.
- Schiestl R, Dominska M, Petes T D. Mol Cell Biol. (1993): 13:2697–2705
- Scheeren-Groot E P, Rodenburg K W, den Dulk-Ras A, Turk S C H J, Hooykaas P J J. J Bacteriol. (1994): 176:6418–6426.
- Turk S C H J, van Lange R P, Sonneveld E, Hooykaas P J J. J Bacteriol. (1993): 175:5706–5709
- Steensma H Y, Holterman L, Dekker I, Van Sluis C A, Wenzel T J. Eur J Biochem. (1990): 191:769–774
- Bevan M. Nucleic Acids Res. (1984): 22:8711–8721.
- Holm C, Meeks-Wagner D W, Fangman W L, Botstein P. Gene. (1986): 42:169–173.
- Gibson S I, Sommerville C. (1992). In: Methods in Arabidopsis Research. Koncz C, Chua N-H, Schell J, editors. Singapore: World Scientific. 119–143.
- de Jonge P, de Jongh F C M, Meijers R, Steensma H Y, Scheffers W A. Yeast. (1986): 2:193–204
- Schwartz D C, Cantor C R. Cell. (1984): 37:67–75
- Tinland B, Schoumacher F, Gloeckler V, Bravo-Angel A M, Hohn B. EMBO J. (1995): 14:3585–3595
- van der Graaf E, den Dulk-Ras
- Roth D B, Wilson J H. Mol Cell Biol. (1986) 6:4295–4304.
- Zamaroczy M, Bernardi G. Gene. (1986) 47:155–177
- Thorsness P E, Fox T D. Nature (London) (1990): 346:376–379
- Dürrenberger F, Crameri A, Hohn B, Koukolíková-Nicola Z. Proc Natl Acad Sci USA. (1989): 86:9154–9158
- Pansegrau W, Schröder W, Lanka E. Proc Natl Acad Sci USA. (1993): 90:2925–2929.
Citation: Hamid Kheyrodin. (2019). Methods of study in Molecular Agrobacterium and genes. Journal of Biotechnology and Immunology 1(1).
Copyright: © 2019 Hamid Kheyrodin. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.


