Research Article
Volume 2 Issue 1 - 2020
In Silico Analysis of a Proposed New Generation Multiepitopic Vaccine Against HIV: Can we Facilitate Immune System Challenges?
Department of Genetics, Faculty of Science, University of Shahrekord, Shahrekord, Iran.
*Corresponding Author: Ali Mohammad Ahadi, Department of Genetics, Faculty of Science, University of Shahrekord, Shahrekord, Iran.
Received: March 16, 2020; Published: March 25, 2020
Abstract
Development of new vaccines against infectious disease is permanent challenge in hygiene organization of every country. AIDS is one of the most dispersed infectious disease word wild. High prevalence of HIV may be because of the low ability of WHO in treatment of this disease. It seem vaccination can be a powerful strategy in this case. However several generation of vaccines introduced in past decade, development of the new effective vaccines is necessary. In this study, we introduced a new strategy in design and development of next generation of vaccines against HIV or other infectious diseases with hard treatment process. Employment of molecular adjuvants can help as a bypass in effective immune response. In this study we applied gp120 protein of HIV joined with CD4 in combined with different linkers. Epitope mapping of our construct confirmed high antigenic properties and effective recognition by B-cell and T-cell receptors.
Keywords: HIV; AIDS; Vaccine; Epitope mapping
Introduction
The HIV is one of the major implications for public health worldwide [1]. After 30 years passed from HIV discovery as the cause of AIDS [2, 3], there are more than 36.7 million people affected by HIV. Moreover, there are about 90% of the infected population living in developing countries [4]. Behalf on unavailability of antiretroviral drugs in these countries, development of new effective HIV-1 vaccine generation would be necessary for the elimination of the AIDS pandemic. Unfortunately, there is no fully effective HIV vaccine till present time [8]. Normally, humoral immunity is the first challenge for vaccine-induced immunity against some infectious diseases, such as AIDS. Many researches have deal with humoral anti-HIV immunity [11]. The extraordinary worldwide genomic and phenotypic variance of HIV-1 is one of the greatest obstacles in development of effective vaccines [14]. Some researchers have showed coincident emergence of CTL responses against virus during acute infection synchronous with the control of primary viremia [21-23]. One of the Viral targeting T lymphocyte responses limitations is the mutagenesis in T cell specific epitopes of virus for evading from cellular immunity [24-26]. It seem the most important limitation in achievement to an effective vaccine with cellular immune responses is non-protectively administration of them against HIV-1 infection and the virus rapidly establishes latent titer [27, 28]. Moreover, it is unclear whether T-cell specific vaccine will be able to function rapidly enough within the first few days in order to induce immunity against acute HIV-1 infection[10]. In this study, in order to overcome these limitations, we are designed a fusion protein containing a human CD4 protein as co-receptor of HIV-1 and HIV-1 envelope glycoprotein. This strategy can help immune system for firing of an effective response against HIV. Application of referable software in our in silico study can ensure this approach.
Material and Methods
Data collection and linker design and modeling
The 3D structure of human CD4 protein (PDB entry1WIP) and HIV-1 gp120 glycoprotein (PDB entry 2nzx) were extracted from RCSB database [29]. According to Ryoichi Arai study at 2001 [30], six helical linkers were arranged and designed. The chosen linkers were developed to achievement an appropriate conformational and dynamical structure. The Servers and the software that are used in this study are listed in Table1. The PHYRE2 server [31] and YASARA software[32] were employed for prediction of 3D structure of linkers. Our proposed fusion proteins were containing three different compartment with different source. We used the Chimera 1.8 software [33] to joining the CD4 to every linker and joining this complex to gp120 glycoprotein in the next step. Physical and chemical properties of the models were analyzed with ProtParam server[34].
The 3D structure of human CD4 protein (PDB entry1WIP) and HIV-1 gp120 glycoprotein (PDB entry 2nzx) were extracted from RCSB database [29]. According to Ryoichi Arai study at 2001 [30], six helical linkers were arranged and designed. The chosen linkers were developed to achievement an appropriate conformational and dynamical structure. The Servers and the software that are used in this study are listed in Table1. The PHYRE2 server [31] and YASARA software[32] were employed for prediction of 3D structure of linkers. Our proposed fusion proteins were containing three different compartment with different source. We used the Chimera 1.8 software [33] to joining the CD4 to every linker and joining this complex to gp120 glycoprotein in the next step. Physical and chemical properties of the models were analyzed with ProtParam server[34].
| The name of sever/software | Application | link Address | Ref. |
| RCSB | Protein Data Bank | http://www.rcsb.org/pdb/home/home.do | [29] |
| PHYRE2 | Protein fold recognition server | www.sbg.bio.ic.ac.uk/phyre2/ | [31] |
| YASARA | molecular-graphics, modeling and -simulation program | http://www.yasara.org/ | [32] |
| Chimera | an extensible visualization system | https://www.cgl.ucsf.edu/chimera/ | [33] |
| ProtParam | computation of various physical and chemical parameters of proteins | http://web.expasy.org/protparam/ | [34] |
| BCPREDS | B-cell epitope prediction server | http://ailab.ist.psu.edu/bcpred/ | [35] |
| ABCpred | predict B cell epitope(s) | http://www.imtech.res.in/raghava/abcpred | [36] |
| Bcepred | Prediction of linear B-cell epitopes | http://www.imtech.res.in/raghava/bcepred/ | [37] |
| Ellipro | Antibody epitope prediction | (http://tools.immuneepitope.org/tools/ElliPro/iedb_input | [38] |
| CTLPred | prediction of CTL epitopes | http://www.imtech.res.in/raghava/ctlpred/ | [39] |
| NetCTL | predicts CTL epitopes | http://www.cbs.dtu.dk/services/NetCTL/ | [40] |
| EpiJen | T cell epitope prediction | www.ddg-pharmfac.net/epijen/ | [41] |
| TAPpred | T cell epitope prediction | http://www.imtech.res.in/raghava/tappred | [42] |
Table 1: The list of server and software utilized in this study.
Prediction of B-cell epitopes
In order to the prediction of B-cell-specific epitopes, primary sequences of the proposed fusion proteins were submitted to 4 servers included; BCPREDS [35], ABCpred [36], Bcepred [37] and Elipro [38]. In addition, Elipro [38] server was utilized to predict conformational epitopes indictable by B lymphocytes.
In order to the prediction of B-cell-specific epitopes, primary sequences of the proposed fusion proteins were submitted to 4 servers included; BCPREDS [35], ABCpred [36], Bcepred [37] and Elipro [38]. In addition, Elipro [38] server was utilized to predict conformational epitopes indictable by B lymphocytes.
Prediction of T-cell epitopes
T-cell-specific epitopes were identified by submission of primary sequences of the proposed fusion proteins, to CTLPred [39], EpiJen [41], TAPpred [42] and NetCTL [40] servers.
T-cell-specific epitopes were identified by submission of primary sequences of the proposed fusion proteins, to CTLPred [39], EpiJen [41], TAPpred [42] and NetCTL [40] servers.
Results
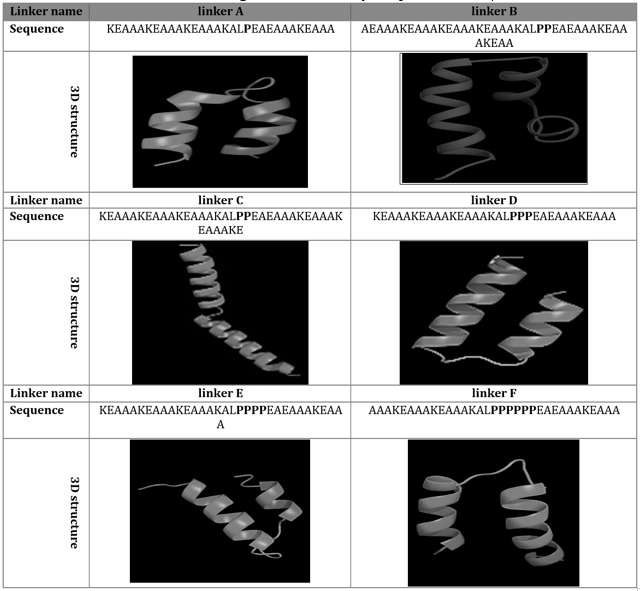
Primary and 3D structure of linkers designed for joining of fusion protein compartments are listed in table 2. As it is shown in 3D structure of every linker, the number of proline amino acid residues can affect the structure effectively. Constructs are named “A” to “F” in this study based on the type of their linker. By utilizing of six linkers, six models were obtained respectively (figure 1).
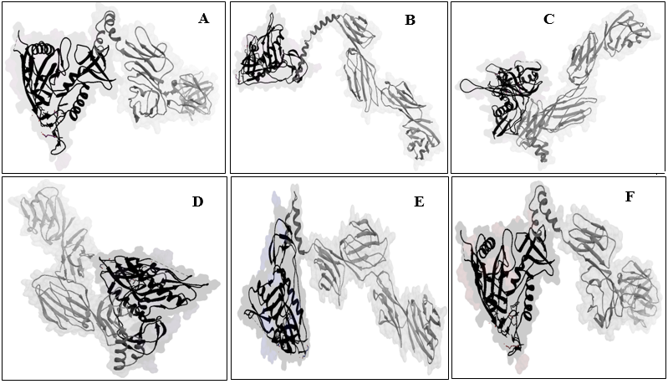
Figure 1: 3D structure of fusion proteins. CD4 part colored by light gray, the linker colored by gray and gp120 colored by black. Name of every model is based on that linker is applied for every construct. Linker must act as a spacer between two macromolecule.
A summary of physical and chemical properties of our proposed fusion proteins extracted from ProtParam server is presented in the table 3. Result showed that all of the presented models are hydrophobic. The Aliphatic index of the models show that all of them have a good thermostability. As it is shown in table 4, the main structure of fusion proteins consist of β-sheet and coil.
| Model | Number of amino acids | Molecular weight (kD) | Theoretical pI | Instability index | Aliphatic index | GRAVY |
| A | 699 | 770.7 | 8.86 | 38.67/stable | 86.05 | -0.293 |
| B | 709 | 779.8 | 8.80 | 38.81/stable | 85.68 | -0.291 |
| C | 707 | 779 | 8.85 | 39.08/stable | 85.5 | 0-.305 |
| D | 701 | 772.1 | 8.86 | 39.13/stable | 85.81 | -0.297 |
| E | 702 | 773.6 | 8.86 | 39.37/stable | 85.68 | -0.299 |
| F | 701 | 771.9 | 8.86 | 40.35/unstable | 85.25 | -0.300 |
Table 3: Physical and chemical properties of the proposed fusion proteins.
| Model | α-Helix (%) | β-Sheet (%) | Turn (%) | Coil (%) |
| A | 10.6 | 40.3 | 8.9 | 40.2 |
| B | 13.5 | 39.8 | 8.2 | 38.5 |
| C | 13.8 | 39.9 | 7.6 | 39.6 |
| D | 12.1 | 40.4 | 7.7 | 39.8 |
| E | 11.4 | 40.2 | 7.7 | 40.7 |
| F | 11.5 | 40.2 | 8.3 | 40 |
Table 4: 2D structure of the proposed fusion proteins.
Different bioinformatics programs were used to predict the linear B-cell epitopes whose results are presented in table 5. In addition to the linear epitopes, discontinuous epitopes were predicted too, as it is shown in tables 6. For each model Elipro server predicted several conformational epitopes but we are chosen only the epitopes with 0.7 and higher score. Also, T lymphocyte specific epitopes were predicted by various programs and the results including the sequence of epitope, the PI of models and GRAVY score of them, are presented in table 7.
| Motif | position (at "A" model) | Position (at "B" model) | Position (at "C" model) | Position (at "D" model) | Position (at "E" model) | Position (at "F" model) | Length | GRAVY Score | PI | Software |
| EVVIRSVNFTDNAKTI | 516-531 | 526-541 | 524-239 | 518-533 | 519-534 | 519-534 | 16 | 0.1 | 6.17 | 1,2 |
| EIFRPGG | 674-680 | 684-690 | 682-688 | 676-682 | 677-683 | 677-683 | 7 | -0.443 | 6.10 | 1,2,3 |
| TIIFKQSSGGD | 576-586 | 586-596 | 584-594 | 578-588 | 579-589 | 579-589 | 11 | -0.200 | 5.5 | 1,2,3 |
| GKAMYAPPISGQIRCS | 639-654 | 649-664 | 647-662 | 641-656 | 642-657 | 642-657 | 16 | -0.113 | 9.31 | 1,2 |
| TWFNSTGS | 612-619 | 622-629 | 620-627 | 614-621 | 615-622 | 615-622 | 8 | -0.625 | 5.19 | 1,2,3 |
| VVLVNVTENFNMWKND | 395-410 | 405-420 | 403-418 | 397-412 | 398-413 | 398-413 | 16 | -0.075 | 4.37 | 1,2,4 |
| PIHYCAPAGFAI | 461-472 | 471-482 | 469-480 | 463-474 | 469-480 | 469-480 | 12 | 0.967 | 7.12 | 1,2,4 |
| ISLWDQSLKPCVKLTP | 420-435 | 430-445 | 428-443 | 422-437 | 464-475 | 464-475 | 16 | 0.087 | 8.20 | 1,2,4 |
| NGTGPCTNVSTVQC | 481-494 | 491-504 | 489-502 | 483-496 | 484-497 | 484-497 | 12 | -0.117 | 5.51 | 1,2,4 |
| EVWGPTS | 304-310 | 304-310 | 304-310 | 304-310 | 304-310 | 304-310 | 7 | -0.529 | 4.00 | 2,3 |
| CEVEDQK | 84-90 | 84-90 | 84-90 | 84-90 | 84-90 | 84-90 | 7 | -1.600 | 4.14 | 1,2,3,4 |
| TLTLESPPGSSPSVQC | 115-130 | 115-130 | 115-130 | 115-130 | 115-130 | 115-130 | 16 | -.156 | 4.00 | 1,2,3,4 |
| WQAERASSSK | 215-224 | 215-224 | 215-224 | 215-224 | 215-224 | 215-224 | 10 | -1.510 | 8.75 | 1,3,4 |
| QIASKLR | 562-568 | 572-578 | 570-576 | 564-570 | 565-571 | 565-571 | 7 | -0.371 | 11 | 1,2,3, |
| TCTASQKK | 15-22 | 15-22 | 15-22 | 15-22 | 15-22 | 15-22 | 8 | -1.150 | 9.30 | 1,3,4 |
*Servers used for prediction are shown as number described as follows: 1) BCPred 2) ABCpred 3) Bcepred 4) Elipro.
Table 5: B-Cell Linear Motif Prediction of Model A-F.
Table 5: B-Cell Linear Motif Prediction of Model A-F.
| Motif | sequence | Position at " A" model | Position at "B" model | Position at "C" model | Position at "D" model | Position at "E" model | Position at "F" model | Length | GRAVY Score | PI | Software |
| 1 | DTYICEV | 80-86 | 80-86 | 80-86 | 80-86 | 80-86 | 80-86 | 7 | 0.314 | 3.67 | 1,4 |
| 2 | FQKASSI | 179-185 | 179-185 | 179-185 | 179-185 | 179-185 | 179-185 | 7 | 0.014 | 8.75 | 1,4 |
| *3 | KKLPLHLT | 251-258 | 251-258 | 251-258 | 251-258 | 251-258 | 251-258 | 7 | -0.237 | 10.00 | 1,3 |
| 4 | IVQLNTSVE | 532-540 | 542-550 | 540-548 | 534-542 | 535-543 | 535-543 | 9 | 0.522 | 4.00 | 1,2 |
| 5 | GSDTITLPC | 618-626 | 628-636 | 626-634 | 620-628 | 621-629 | 621-629 | 9 | 0.344 | 3.80 | 2,4 |
| *6 | LTANSDTHL | 100-108 | 100-108 | 100-108 | 100-108 | 100-108 | 100-108 | 9 | -0.333 | 5.08 | 2,4 |
| *7 | ASSSKSWIT | 220-229 | 220-229 | 220-229 | 220-229 | 220-229 | 220-229 | 9 | -0.267 | 8.80 | 2,4 |
| 8 | LLSDSGQVL | 346-354 | 346-354 | 346-354 | 346-354 | 346-354 | 346-354 | 9 | 0.733 | 3.80 | 1,2,4 |
| *9 | AILKCNNKT | 471-479 | 481-489 | 479-487 | 473-481 | 474-482 | 474-482 | 9 | -0.322 | 9.32 | 2 |
| *10 | ARAKWNNTL | 552-560 | 562-570 | 560-568 | 554-562 | 555-563 | 555-563 | 9 | -1.067 | 11.00 | 3 |
| 11 | FTDNAKTII | 524-532 | 534-545 | 532-540 | 526-534 | 527-535 | 527-535 | 9 | 0.144 | 5.84 | 4 |
| 12 | LSVSQLELQ | 144-152 | 144-152 | 144-152 | 144-152 | 144-152 | 144-152 | 9 | 0.389 | 4.00 | 1 |
*Antigenic motif were identified by various server. Servers used for prediction are shown as number described as follows:
1) CTLpred 2) Epijene 3) Tappred 4) NetCTL. Hydrophilic motifs are marked with an asterisk.
Table 7: T cell Motif Prediction of Model A-F.
1) CTLpred 2) Epijene 3) Tappred 4) NetCTL. Hydrophilic motifs are marked with an asterisk.
Table 7: T cell Motif Prediction of Model A-F.
Discussion
We are presented in this article, a new insight applicable in design of more effective vaccines against HIV. HIV was detected in 2.1 million people in 2015 and 1.1 million affected individuals were died by it [4]. Nowadays, very active antiviral reagent that targets replication of virions are available [5-7] but HIV-1 has not been removed from populations [8]. An HIV-1 vaccine must be can either to prevent infection and to reduce viral titer and progression of disease. A powerful vaccine would completely inhibit infection and provide stable immunity against HIV-1. In spite of different challenges, this degree of protection is not even achieved[10]. In these situations, improvement of vaccination strategies is necessary. Our study focuses on this weak point.
In several study, it is used used monomeric HIV-1 Env gp120 protein to induce specific humoral immune responses against viral envelop protein [12, 13]. If we consider high diversity of HIV-1, we will therefore need an effective immunogenic vaccine to promote immune system responses against different variants of HIV. We have considered this properties in our proposed vaccine. We analyzed immunogenicity of proposed fusion peptide in front of TCR and BCR. Our results are supported by other study that have been reported cross-reactive humoral and cellular immune responses against conserved regions of the virus [10].
There is a high variance in the first structure of immunogenic proteins of HIV. For example, Env protein of virus has up to 35% difference in amino acid sequence in variants [14, 15]. On the other hand, N-glycosylation of Env glycoprotein can hide some epitopes from immune system recognition result in decreasing of efficient humoral immunity [16, 17].
We proposed an effective method for designing of new vaccines that can facilitate action of Antigen presentation cells (APC). In our plan, a proline amino acid residue linker, joint gp120 protein to CD4 receptor in a “U” shaped manner. We are believing this construct can present gp120 beside of CD4 to the effector cells such as B and Ts cells. Table 1-4 and figure-1 shows conformational shape and some properties of our proposed construct. These data confirm a stable shape of final construct with highly stable secondary structures (table-4). In addition table 3 show a basic PI for all of models and instability index lower than 40 for A-E models that means optimal stability of these constructs. On the other hand, aliphatic index and GRAVY confirmed two parameters necessary for action of a molecule as an effective immunogenic particle.
Our proposed gp120-linker-CD4 construct can be supported by some reported biophysical studies. It was explained in some researches that highly immunogenic variable motives elicit type-specific antibodies that may redirect humoral responses away from conserved regions. In addition, critical changes in order to appearance of conserved motives happens after the binding of Env to its cellular receptor CD4 that result in an extensive conformational change [18]. We analyzed immunogenic parameters of new constructs by utilizing of different well known software and servers.
B-Cell Linear motives in the models A-F, showed a GRAVY score equal with .961 in minimum and maximum equal with -1.561 that confirm a good condition for an immune response(table 5). Conformational B-cell epitopes identified by Ellipro software. All of proposed models have an Ellipro score more than 0.7 that is indication of high immunogenic properties (table 6). T-cell specific epitopes are showed in table 7. GRAVY score of some motives in range of TCR detection that means we can expect an effective immune cellular response.
In summary, we introduced in this paper an new strategy can help to immune system in order to direct detection of vaccine protein independently of interaction between antigen and CD4 receptor. Bioinformatics analyses of proposal protein confirmed it as an effective candidate vaccine against HIV. However construction of this fusion protein by using of recombinant DNA technology and animal challenges experiments will help us for a better view.
Acknowledgments: We thanks Shahrekord University for its financial supports.
References
- Ensoli, B., et al., (2014). Challenges in HIV Vaccine Research for Treatment and Prevention. Front Immunol,. 5: p. 417.
- Barre-Sinoussi, F., et al., (1983). Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS). Science. 220(4599): p. 868-71.
- Gallo, R.C., et al., (1983). Isolation of human T-cell leukemia virus in acquired immune deficiency syndrome (AIDS). Science. 220(4599): p. 865-7.
- WHO, Data and Statistics. http://www.who.int/hiv/data/en/. 2016.
- Walensky, R.P., et al. (2006). The survival benefits of AIDS treatment in the United States. J Infect Dis. 194(1): p. 11-9.
- Palella, F.J., Jr., et al. (1998). Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. N Engl J Med. 338 (13): p. 853-60.
- Ho, D.D., et al. (1995). Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature,. 373(6510): p. 123-6.
- Wang, H.B., Q.H. Mo, and Z. Yang. (2015). HIV vaccine research: the challenge and the way forward. J Immunol Res,. 2015: p. 503978.
- Richman, D.D., et al., (2009). The challenge of finding a cure for HIV infection. Science. 323(5919): p. 1304-7.
- Barouch, D.H. (2008). Challenges in the development of an HIV-1 vaccine. Nature. 455(7213): p. 613-9.
- Miedema, F., (2008). A brief history of HIV vaccine research: stepping back to the drawing board? AIDS. 22(14): p. 1699-703.
- Pitisuttithum, P., et al. (2006). Randomized, double-blind, placebo-controlled efficacy trial of a bivalent recombinant glycoprotein 120 HIV-1 vaccine among injection drug users in Bangkok, Thailand. J Infect Dis. 194(12): p. 1661-71.
- Flynn, N.M., et al. (2005). Placebo-controlled phase 3 trial of a recombinant glycoprotein 120 vaccine to prevent HIV-1 infection. J Infect Dis. 191(5): p. 654-65.
- Gaschen, B., et al. (2002). Diversity considerations in HIV-1 vaccine selection. Science. 296(5577): p. 2354-60.
- Walker, B.D. and B.T. Korber. (2001). Immune control of HIV: the obstacles of HLA and viral diversity. Nat Immunol. 2(6): p. 473-5.
- Wyatt, R., et al., (1998). The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 393(6686): p. 705-11.
- Kwong, P.D., et al., (1998). Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 393(6686): p. 648-59.
- Chen, B., et al., (2005). Structure of an unliganded simian immunodeficiency virus gp120 core. Nature. 433(7028): p. 834-41.
- Richman, D.D., et al., (2003). Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci U S A,. 100(7): p. 4144-9.
- Wei, X., et al., (2003). Antibody neutralization and escape by HIV-1. Nature,. 422(6929): p. 307-12.
- Borrow, P., et al., (1994). Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J Virol,. 68(9): p. 6103-10.
- Koup, R.A., et al., (1994). Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J Virol,. 68(7): p. 4650-5.
- Pantaleo, G., et al., (1994). Major expansion of CD8+ T cells with a predominant V beta usage during the primary immune response to HIV. Nature,. 370(6489): p. 463-7.
- Allen, T.M., et al., (2000). Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature,. 407(6802): p. 386-90.
- Barouch, D.H., et al., (2002). Eventual AIDS vaccine failure in a rhesus monkey by viral escape from cytotoxic T lymphocytes. Nature,. 415(6869): p. 335-9.
- Phillips, R.E., et al., (1991). Human immunodeficiency virus genetic variation that can escape cytotoxic T cell recognition. Nature,. 354(6353): p. 453-9.
- Chun, T.W., et al., (1997). Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature,. 387(6629): p. 183-8.
- Chun, T.W., et al., (1998). Early establishment of a pool of latently infected, resting CD4(+) T cells during primary HIV-1 infection. Proc Natl Acad Sci U S A,. 95(15): p. 8869-73.
- Berman, H.M., et al., (2000). The Protein Data Bank. Nucleic Acids Res,. 28(1): p. 235-42.
- Arai, R., et al., (2001). Design of the linkers which effectively separate domains of a bifunctional fusion protein. Protein Eng,. 14(8): p. 529-32.
- Kelley, L.A., et al., (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protocols,. 10(6): p. 845-858.
- Krieger E, Vriend G. (2014). YASARA View - molecular graphics for all devices - from smartphones to workstations. Bioinformatics. Oct 15;30(20):2981-2.
- Pettersen, E.F., et al., (2004). UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem,. 25(13): p. 1605-12.
- Walker, J.M., (2005). The Proteomics Protocols Handbook.: Humana Press.
- El-Manzalawy, Y., D. Dobbs, and V. Honavar, (2008). Predicting linear B-cell epitopes using string kernels. J Mol Recognit,. 21(4): p. 243-55.
- Saha, S. and G.P. Raghava. (2006). Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins,. 65(1): p. 40-8.
- Saha S, Raghava GPS. (2004). BcePred: Prediction of continuous B-cell epitopes in antigenic sequences using physicochemical properties. In ICARIS,. 3239: p. 197-204.
- Ponomarenko, J., et al., (2008). ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinformatics,. 9: p. 514.
- Bhasin, M. and G.P. Raghava. (2004). Prediction of CTL epitopes using QM, SVM and ANN techniques. Vaccine,. 22(23-24): p. 3195-204.
- Larsen, M.V., et al., (2007). Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinformatics,. 8: p. 424.
- Doytchinova, I.A., P. Guan, and D.R. Flower. (2006). EpiJen: a server for multistep T cell epitope prediction. BMC Bioinformatics,. 7: p. 131.
- Bhasin, M. and G.P. Raghava. (2004). Analysis and prediction of affinity of TAP binding peptides using cascade SVM. Protein Sci,. 13(3): p. 596-607.
Citation: Atefeh Amiri, Ali Mohammad Ahadi, Hoda Ayat. (2020). In Silico Analysis of a Proposed New Generation Multiepitopic Vaccine Against HIV: Can we Facilitate Immune System Challenges? Journal of Biotechnology and Immunology 2(1).
Copyright: © 2020 Ali Mohammad Ahadi. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.