Research Article
Volume 5 Issue 1 - 2023
Complete Genome Sequences of Citrus Yellow Mosaic Virus Infecting Different Citrus Species from India
1Plant Virology Unit, Division of Plant Pathology, Indian Agricultural Research Institute New Delhi- 110012, India
2PC Unit, Sesame and Niger (ICAR) Jawaharlal Nehru Krishi Vishwa Vidyalaya, Jabalpur 482 004, Madhya Pradesh, India
2PC Unit, Sesame and Niger (ICAR) Jawaharlal Nehru Krishi Vishwa Vidyalaya, Jabalpur 482 004, Madhya Pradesh, India
*Corresponding Author: Kailash N Gupta, Plant Virology Unit, Division of Plant Pathology, Indian Agricultural Research Institute New Delhi- 110012, India.
Received: January 10, 2023; Published: February 03, 2023
Abstract
A study was undertaken to find out the genomic variability in Citrus yellow mosaic virus (CMBV), a bacilliorm virus under the genus Badnavirus, infecting different citrus species in India and their phylogenetic relationship with other badnaviruses. Comparison of genome sequences (CMBVSON: Acc. No FJ617224) of four isolates CMBVRL, Acc. No DQ875213; CMBVPM, Acc. No EU489745;CMBVAL1, Acc. No EU7081317; CMBVAL 2, Acc. No EU489744;CMBVSOP, Acc. No EU489744) of CMBV infecting different species of citrus with previously sequenced three CMBV isolates indicated variability in coding region of ORFs 1, 2 and 3 of all the CMBV isolates infecting same or different citrus species with highest variability in coding region of ORF3. Coding region of ORF 4, 5 were also highly variable in CMBV isolates but they were highly conserved in CMBV isolates infecting Acid lime, ORF 6 was comparatively conserved and was identical in CMBV isolate infecting Acid lime. All the CMBV isolates shared maximum identity with Cacao swollen shoot virus (CSSV) in ORF 1 and 3 indicating that CMBV isolates are more closely related to CSSV than other badnaviruses. This study has implication in determining the diversity and diagnosis of CMBV
Introduction
Citrus mosaic is a widely distributed disease of citrus in India particularly of sweet oranges [Citrus sinensis (L) Osbeck] and pummelo [Citrus grandis (L) Osbeck] [1, 2]. The disease was shown to be graft transmitted to 13 citrus species and their cultivars [2]. A mealybug Planococcus citri was established as the vector of the disease [9]. It is caused by Citrus yellow mosaic virus (CMBV) a bacilliform non-enveloped DNA virus grouped under the genus Badnavirus of family Caulimoviridae [1, 4). The complete genome of CMBV was sequenced and it consisted of 7559 bp and 6 ORFs. [7]. Here, we report the complete genome sequences of CMBV isolates infecting sathgudi sweet orange (Citrus sinensis (L) Osbeck), acid lime (Citrus aurantifolia (Christm.) swing, Rangpur lime (Citrus limonia (Osb).) and its comparison with 3 more isolates of CMBV and other badnaviruses.
Methods and Material
Provenance of the virus: The CMBV isolates infecting different citrus species vizs., of Sathgudi sweet orange (Citrus sinensis (L.) Osbeck), acid lime (Citrus aurantifolia (Christm) swing, Rangpur lime (Citrus limonia (Osb) were collected from different locations viz., Nagri (Chittor District), Pulvendela (Cudappa, District) and Tirupathi District, of Andhra Pradesh southern part of India. Total DNA was extracted from infected citrus tissues using DNeasy Plant Mini Kit (Qiagen, Germany) according to the manufacture’s instructions. The genome fragments of 4 isolates of CMBV were PCR amplified using total DNA and the genome sequences were determined using eight sets of overlapping primers. The full genome sequences were CMBV compared with sequences of 3 other isolates of CMBV (Acc. Nos, AF347695; EU489744; EU489745) and 14 other badnaviruses available in Gene bank (Table 1). Nucleotide and amino acid sequence data were analyzed using the BioEdit sequence alignment editor version 5.09.04 [6]. Sequence phylograms were constructed using the software Clustal W [10].
Sequence Properties
The complete genome of CMBV sweet orange Nagri (CMBVSON) and CMBV sweet orange Pulvendual (CMBVSOP) are 7558 and 7497 nucleotides long respectively while that of CMBV acid lime Tirupathi (CMBVAL) and CMBV Rangpurlime Tirupathi (CMBVRL) are 7498 and 7522 nucleotides long respectively. The genome sequence of 4 CMBV isolates have non coding region of ranging from 717 to 731 bp. The sequence has been deposited in GenBank (CMBVSOP: Acc. No. EU708316, CMBVAL: Acc. No. EU 7081317, CMBVRL: Acc. No. DQ875213). The complete genome of 3 other isolates of CMBV infecting sweet orange, acid lime and Pummelo (Acc No. AF347695; EU489744, EU489745) were 7559, 7437 and 7487 nt. with non coding region varying from 720 to 731.
The complete genome of CMBV sweet orange Nagri (CMBVSON) and CMBV sweet orange Pulvendual (CMBVSOP) are 7558 and 7497 nucleotides long respectively while that of CMBV acid lime Tirupathi (CMBVAL) and CMBV Rangpurlime Tirupathi (CMBVRL) are 7498 and 7522 nucleotides long respectively. The genome sequence of 4 CMBV isolates have non coding region of ranging from 717 to 731 bp. The sequence has been deposited in GenBank (CMBVSOP: Acc. No. EU708316, CMBVAL: Acc. No. EU 7081317, CMBVRL: Acc. No. DQ875213). The complete genome of 3 other isolates of CMBV infecting sweet orange, acid lime and Pummelo (Acc No. AF347695; EU489744, EU489745) were 7559, 7437 and 7487 nt. with non coding region varying from 720 to 731.
Result and Discussion
Location, number of ORFS, comparison of ORF and phylogenetic analysis of Citrus yellow mosaic virus of different citrus species and other badnaviruses
All six ORFs are present on plus strand. ORF 1 consisted of 143 amino acids in all the CMBV isolates. It showed identity in the range of 96.5% to 96.6%. ORF 2 consisted of 137 amino acids in all the CMBV isolates except CMBVRL in which ORF 2 had 138 amino acid and showed identity in the range of 89.1% to 99.2%. Finding of ORF 1 and 2 are not known. Yet ORF 3 is the largest ORF and has variable size in different isolates.
All six ORFs are present on plus strand. ORF 1 consisted of 143 amino acids in all the CMBV isolates. It showed identity in the range of 96.5% to 96.6%. ORF 2 consisted of 137 amino acids in all the CMBV isolates except CMBVRL in which ORF 2 had 138 amino acid and showed identity in the range of 89.1% to 99.2%. Finding of ORF 1 and 2 are not known. Yet ORF 3 is the largest ORF and has variable size in different isolates.
It codes for polyproteins such as movement protein, coat protein, aspartic protease, RNase H and reverse transcriptase [7]. The number of amino acid in ORF 3 varies from 1744 to 1983 and it shows identity in the range of 84.2% to 87.1%. ORF 4, ORF 5 and ORF 6 overlaps with ORF 3. Function of ORF 5 and ORF 6 are also not known. ORF 4 overlaps with ORF 3 in its N terminal end. ORF 4 is of highly variable in different CMBV isolates and amino acids number varies from 85 to 181. It showed identity in the range of 28.4% to 87.1%. ORF 5 in different isolates consists of 95 to 103 amino acid and showed identity in the range of 71.8 % to 95.1 % in different isolates. ORF 6 was relatively conserved and had 154 amino acids in all isolates except CMBVSON where it has 153 amino acids and identity in ORF 6 varies from 93.5% to 96.1% in different CMBV isolates. Comparative analysis revealed that the different CMBV isolates shared 87.3% to 98.9% identity in nucleotide of their genome (Table 1). A TATATAA box was present in the intergenic region. Upstream of the TATA box were CACAAT and TGACG sequences similar to that found upstream of the TATA box in the 35S promoter of cauliflower mosaic virus (CaMV) [ 8]. Downstream of the putative CMBV promoter was a possible polyadenylation signal (AATAAA) [3] similar to that previously reported in CMBV and also seen in other badnaviruses. [5]. Most of the badnaviruses have three ORFs except CMBV (6 ORFs), Cacao swollen shoot virus (CSSV, 5 ORFs), Dracaena mottle virus (DrMV, 7 ORFs) and Dioscorea bacilliform virus (DaBV, 4 ORFs). Comparative sequence analysis with other badnaviruses showed that CSSV, DaBV and DrMV had maximum identity of 44.1% to 48.3% with CMBV isolates in complete genome sequence but DaBV had maximum identity with CMBV isolates (47.9%) but DrMV had 6 ORFs, the only other badnaviruses having 6 ORFs like CMBV, DaBV and DrMV showed more identity in ORF II compared to CSSV. The study reveals complex evolution of badnaviruses in plants.
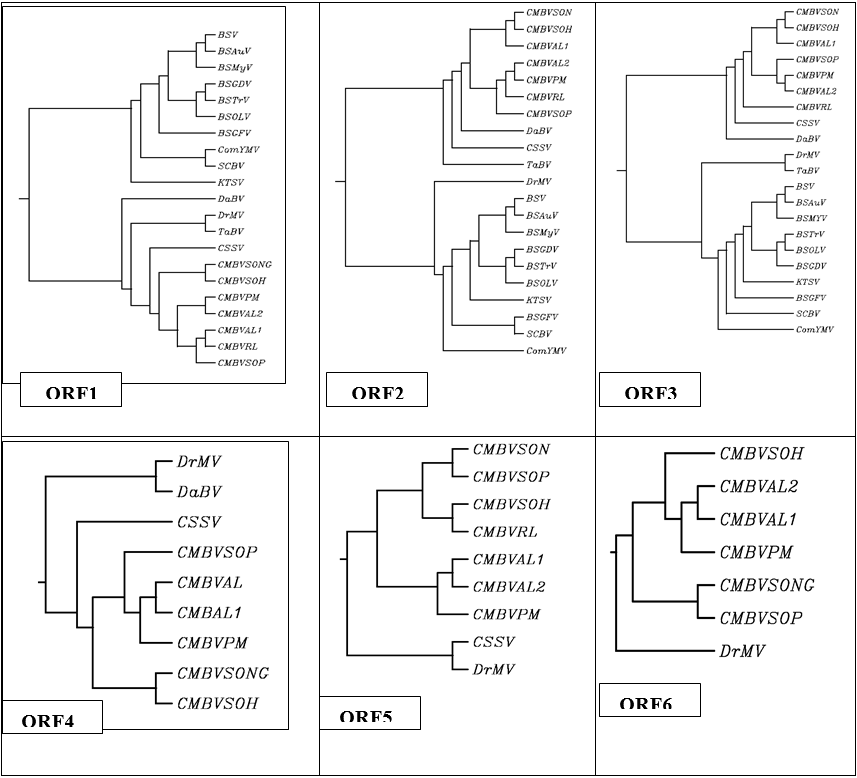
Figure 1: Cluster dendogram showing the relationships between the deduced amino acid sequences of the complete genome of Cirus yellow mosaic virus from India with those of known CMBV sequence and other Badnavirus from GeneBank. The Phylogenetic tree was constructed using Clustal W. Accession number for compared CMBV and other badnavirus sequences; Banana streak virus Acuminata (BSAuV) China , DQ092436; Banana streak virus (BSV) NC_008018; Banana streak virus Tr (BSTrV), India , DQ859899; Banana streak virus GD (BSGDV), China , DQ451009; Banana streak virus (BSOLV), USA, NC_003381 ; Banana streak Mysore virus (BSMyV), USA, DQ092436 ; Banana streak GF virus (BSGFV), USA, NC_007002; Cacao swollen shoot virus (CSSV), USA; NC001547; Citrus yellow mosaic virus (CMBVSONG) CMBVSON: Acc. No FJ617224 this studies); Citrus yellow mosaic virus(CMBVH) ,USA ,AF347695; Citrus yellow mosaic virus RL (CMBVRL), India, DQ875213; Citrus yellow mosaic virus PM (CMBVPM), EU489745; Citrus yellow mosaic virus AL(CMBVAL2), EU489744; Citrus yellow mosaic virus AL ( CMBVAL1) ,EU7081317; Citrus mosaic virus SOP (CMBVSOP) EI708316; Commelina yellow mottle virus (ComYMV, USA, X52938; Dioscorea bacilliform virus (DaBV), France, DQ822074; Dracaena mottle virus (DrMV), China, DQ473478; Kalanchoe top-spotting Kalanchoe top-spotting virus (KTSV), USA, NC-004540; Sugarcane bacilliform virus (SCBV), USA, NC-003031; Taro bacilliform virus (TaBV), Australia; AF357836.
| Viruses | Nucleotide Sequence Identity range % |
ORFI | ORFII | ORFIII | ORFIV | ORFV | ORFVI | ||||||
| CMBV Isolates | Size (aa) | Identity Range % | Size (aa) | Identity range % |
Size (aa) | Identity Range % | Size (aa) | Identity range % |
Size (aa) | Identity range % |
Size | Identity Range % | |
| CMBVSONG | 87.4 - 96.0 | 143 | 95.1--98.6 | 137 | 89.1--99.2 | 1744 | 87.1-84.9 | 164 | 50.5-87.1 | 103 | 69.5-95.1 | 153 | 92.8-96.1 |
| CMMVSOH | 87.3 - 96.0 | 143 | 93.7-- 98.6 | 137 | 89.8--99.2 | 1983 | 87.1-95.9 | 178 | 34.6-87.1 | 95 | 71.1-75.7 | 154 | 94.1-97.4 |
| CMBVSOP | 88.1 - 90.2 | 143 | 95.1--98.6 | 137 | 89.8--92.7 | 1967 | 83.7-93.8 | 85 | 28.4-38.6 | 103 | 71.4-95.1 | 154 | 94.1-95.4 |
| CMBVAL1 | 88.8 - 91.7 | 143 | 95.1--99.3 | 137 | 89.7--91.9 | 1968 | 84.9 -95.9 | 89 | 55.8 - 98.3 | 95 | 71.8-96.9 | 154 | 93.5-98.7 |
| CMBVRL | 88.6 - 91.6 | 143 | 95.8--99.3 | 138 | 89.1--93.4 | 1954 | 83.4-94.3 | 148 | 29.9-55.8 | 95 | 69.0-90.5 | 154 | 95.4-96.1 |
| CMBVAL2 | 88.0 - 98.9 | 143 | 95.1--98.6 | 137 | 90.5--97.8 | 1976 | 84.4-98.6 | 181 | 38.6-61.3 | 95 | 71.8-96.9 | 154 | 95.1-98.7 |
| CMBVPM | 87.4 - 98.9 | 143 | 94.4--98.6 | 137 | 89.7--97.8 | 1979 | 84.2-98.6 | 181 | 37.5-98.3 | 97 | 69.5-96.9 | 154 | 92.8-98.7 |
| *Other Badnaviruses | |||||||||||||
| CSSV | 44.1 - 44.7 | 143 | 53.8--55.9 | 145 | 19.0--21.0 | 1816 | 47.8-50.3 | 113 | 03.0- 07.2 | A | 6.8-9.1 | A | - |
| BSV | 34.6 - 35.5 | 176 | 24.4--25.0 | 132 | 19.1--21.2 | 1900 | 31.3-33.3 | A | - | A | - | A | - |
| BSAuV | 34.6 - 35.5 | 176 | 24.4 25.0 | 132 | 19.1--21.2 | 1900 | 31.3--33.3 | A | - | A | - | A | - |
| BSTrV | 32.2--33.6 | 176 | 27.1--27.6 | 134 | 17.5--20.2 | 1709 | 33.4-33.7 | A | - | A | - | A | - |
| BSGDV | 32.9--33.2 | 177 | 27.1 --28.2 | 134 | 17.5--20.0 | 1709 | 33.3-33.8 | A | - | A | - | A | - |
| BSGFV | 34.9--35.0 | 177 | 21.3-21.9 | 134 | 15.4--17.7 | 1832 | 31.3-33.8 | A | - | A | - | A | - |
| BSMyVV | 35.0 --35.2 | 176 | 24.4--25.0 | 132 | 19.1--21.2 | 1900 | 31.3-33.3 | A | - | A | - | A | - |
| BSOLV | 34.1 --34.8 | 175 | 27.1--28.2 | 112 | 17.5--20.2 | 1832 | 31.2-33.3 | A | - | A | - | A | - |
| SCBV | 33.2--33.6 | 185 | 18.6--19.1 | 123 | 19.1--24.8 | 1912 | 27.1-29.4 | A | - | A | - | A | - |
| TaBV | 36.5--36.9 | 146 | 33.3--34.6 | 144 | 17.3--18.6 | 1881 | 30.8-33.9 | A | - | A | - | A | - |
| KTSV | 34.4 -35.1 | 173 | 18.1-- 18.7 | 124 | 21.3--23.4 | 1941 | 30.0-32.4 | A | - | A | - | A | - |
| ComYMV | 35.1-35.7 | 200 | 17.5--18.0 | 135 | 14.3--16.3 | 1886 | 32.2-32.7 | A | - | A | - | A | - |
| DaBV | 47.9-48.3 | 143 | 33.7--34.4 | 125 | 31.8--34.7 | 1295 | 25.0-27.8 | 59 | 1.8-.5 | A | - | A | - |
| DrMV | 46.0-46.6 | 149 | 34.6--35.3 | 131 | 26.2--28.5 | 1916 | 34.4--37.3 | 103 | 5.1-12.1 | 91 | 6.8-13.8 | 139 | 22.0-22.7 |
Table 1: Nucleotide sequence and Amino acid percent similarity of different ORFs of CMBVSOH with Badnaviruses.
A-Absent, ND-Not De fined; *Other Badnavirus showing rang of identity with CMBV isolates
Accession number for compared CMBV and other badnavirus sequences vizs., Banana streak virus Acuminata (BSAuV),DQ092436;Banana streak virus (BSV), NC_008018; Banana streak virus TrV (BSTrV), DQ859899; Banana streak GD virus (BSGDV), DQ451009; Banana streak virus OL (BSOLV), NC_003381 ; Banana streak Mysore virus (BSMyV), DQ092436 ; Banana streak GF virus (BSGFV), NC_007002; Cacao swollen shoot virus(CSSV), NC001547; Citrus yellow mosaic virus (CMBVSONG)(this studies); Citrus yellow mosaic virus (CMBVH), AF347695; Citrus yellow mosaic virus RL (CMBVRL), DQ875213; Citrus yellow mosaic virus PM (CMBVPM), EU489745; Citrus yellow mosaic virus AL 2(CMBVAL 2), EU489744; Citrus yellow mosaic virus AL1 (CMBVAL1), EU7081317; Citrus mosaic virus SOP (CMBVSO) , EU489744; Commelina yellow mottle virus (ComYMV), X52938; Dioscorea bacilliform virus (DaBV), DQ822074; Dracaena mottle virus (DrMV), DQ473478; Kalanchoe top-spotting virus (KTSV),NC-004540; Sugarcane bacilliform virus (SCBV), NC-003031; Taro bacilliform virus (TaBV), AF357836
Accession number for compared CMBV and other badnavirus sequences vizs., Banana streak virus Acuminata (BSAuV),DQ092436;Banana streak virus (BSV), NC_008018; Banana streak virus TrV (BSTrV), DQ859899; Banana streak GD virus (BSGDV), DQ451009; Banana streak virus OL (BSOLV), NC_003381 ; Banana streak Mysore virus (BSMyV), DQ092436 ; Banana streak GF virus (BSGFV), NC_007002; Cacao swollen shoot virus(CSSV), NC001547; Citrus yellow mosaic virus (CMBVSONG)(this studies); Citrus yellow mosaic virus (CMBVH), AF347695; Citrus yellow mosaic virus RL (CMBVRL), DQ875213; Citrus yellow mosaic virus PM (CMBVPM), EU489745; Citrus yellow mosaic virus AL 2(CMBVAL 2), EU489744; Citrus yellow mosaic virus AL1 (CMBVAL1), EU7081317; Citrus mosaic virus SOP (CMBVSO) , EU489744; Commelina yellow mottle virus (ComYMV), X52938; Dioscorea bacilliform virus (DaBV), DQ822074; Dracaena mottle virus (DrMV), DQ473478; Kalanchoe top-spotting virus (KTSV),NC-004540; Sugarcane bacilliform virus (SCBV), NC-003031; Taro bacilliform virus (TaBV), AF357836
Conclusion
All the CMBV isolates shared maximum identity with Cacao swollen shoot virus (CSSV) in ORF 1 and 3 indicating that CMBV isolates are more closely related to CSSV than other badnaviruses. This study has implication in determining the diversity and diagnosis of CMBV
Acknowledgement
Authors Profoundly acknowledgement for DBT, GOI and Head, Division of Plant Pathology, Indian Agricultural Research Institute New Delhi- 110012, India.
Authors Profoundly acknowledgement for DBT, GOI and Head, Division of Plant Pathology, Indian Agricultural Research Institute New Delhi- 110012, India.
References
- Ahlawat YS, Pant, RP. Lockhart, B E L,Srivastava M, Chakraborty, NK. Varma, A. (1996). Association of a badnavirus with citrus mosaic disease in India. Plant Disease.80: 590-592.
- Ahlawat YS. (1997). Viruses, greening bacterium and Viroids associated with citrus (Citrus species) decline in India. Indain J Agricultural Sciences. 67: 51-57.
- Boeke JD, Corces VG. (1989). Transcription and reverse transcription of retro-transposons. Annual Review of Microbiology. 43: 403-434.
- Fauquet CM, Mayo MA, Maniloff J, Desselberg U and Ball, LA. (2000). Virus Taxonomy: VIIIth Report of the International Committee on Taxonomy of Viruses. p.800.
- Geering, ADW, McMichael LA, Dietzgen RG, Thomas JE. (2000). Genetic diversity among banana streak virus isolates from Australia. Phytopathology. 90: 921-927.
- Hall TA. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. 41: 95-98.
- Huang, Q. and Hartung, J.S. (2001). Cloning and sequence of an infectious clone of citrus yellow mosaic virus that can infect sweet orange via Agrobacterium mediated inoculation. Journal of General Virology. 82: 2549-2558.
- Odell JT, Nagy F, and Nam HC. (1985). Identification of DNA sequences required for the activity of the mosaic virus 35s promoter. Nature. 313: 810-812.
- Reddy BVB. (1997). Characterization of citrus mosaic virus and to develop methods for its quick detection. Ph.D. Thesis, Indian Agricultural Research Institute.
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W. (1994). Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties, and weight matrix choice. Nucleic Acids Research. 22: 4673-4680.
Citation: K. N. Gupta and V. K. Baranwal. (2023). Complete Genome Sequences of Citrus Yellow Mosaic Virus Infecting Different Citrus Species from India. Journal of Agriculture and Aquaculture 5(1).
Copyright: © 2023 Kailash N Gupta. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
