Research Article
Volume 1 Issue 1 - 2019
Isolation and Identification of Escherichia coli from Dairy Cow Raw Milk in Bishoftu Town, Central Ethiopia
1West Shoa zone, Livestock and fishery resource development office, Dendi district, Ethiopia
2School of Veterinary Medicine, Wolaita Sodo University, Ethiopia
2School of Veterinary Medicine, Wolaita Sodo University, Ethiopia
*Corresponding Author: Haben Fesseha, Wolaita Sodo University, School of Veterinary Medicine, P. O. Box 138, Wolaita Sodo, Ethiopia.
Received: December 16, 2019; Published: December 27, 2019
Abstract
A cross-sectional study was carried out from January to April 2018, to determine the presence of Escherichia coli in raw milk from in dairy farms of Bishoftu. A total of 50 samples were collected from dairy cow and processed bacteriologically and the isolate were tested with a number of biochemical tests for confirmation and identification of E.coli. The study revealed that 42% of the collected raw milk were contaminated with E. coli. Besides, all E. coli isolates exhibited bright pink color with lactose fermentation on MacConkey agar plates, metallic sheen on Eosin Methylene Blue agar plate and gram-negative, pink-colored, small rod-shaped organisms arranged in single with pairs or short chains on Gram’s staining. In this study, there was significat (p< 0.05) association between the body condition, age, and breed of the cow with the isolates of E. coli. In conclusion, the presence and consumption of raw milk may constitute a public health hazard and reduced milk quality due to E. coli. Thus, health professionals should create awearness about milk handling practice, storage and milking process to Dairy farmer and milk collectors.
Key words: Bishoftu; Dairy Cow; Escherichia coli; Raw milk
Introduction
Milk is considered a complete and nutritious food for the new-born mammal and human beings, but it is considered as a good medium for many microorganisms (Leedom, 2006). Raw untreated milk is still used by large number of farm families and workers and by a growing segment of the general population who believe that the milk is not only safe but also imparts beneficial health effects that are destroyed by pasteurization (LeJeune and Rajala-Schultz, 2009). For this reason, utilization of both raw untreated milk and raw milk cheeses has frequently been associated with food-borne illness. Especially, developing countries are mostly affected by food-borne infections (Carbas et al., 2012; Bedasa et al., 2018) because of the prevailing poor food handling and sanitation practices, inadequate food safety regulatory systems, lack of financial resources to invest in safer equipment, and lack of education for food-handlers (FAO and WHO, 2004; Oliver, 2005).
The consumption of raw milk and its products is common in Ethiopia, which lead to the transmission of various diseases (Shunda et al., 2013). The ability of raw milk to support the growth of several pathogenic microorganisms that can lead to spoilage of the product and infections in consumers (Yilma and Faye, 2006; Abera, 2008). Numerous epidemiological reports have implicated raw milk is usually colonized by a variety of zoonotic foodborne pathogens such as Campylobacter jejuni, enterohaemorragic Escherichia coli, Salmonella typhimurium, Listeria monocytogenes, Staphylococcus aureus and Yersinia enterocolitica. These pathogens have been originated from environment in the farm, mixing clean milk with mastitis milk, manure, soil, and contaminated water (Gwida and EL-Gohary, 2013; Nigatu et al., 2017).
Among the major infectious agents, E. coli has been associated with milk and some of dairy products (Abebe et al., 2014; Bedasa et al., 2018). E. coli is Gram-negative, facultative anaerobic, rod-shaped and highly motile bacteria that belong to the family Enterobacteriaceae, and a normal inhabitant of the intestines of animals and humans (Tchaptchet and Hansen, 2011, Virpari et al., 2013, Asmelash, 2015) but its recovery from food may be of public health concern due to the possible presence of enteropathogenic and/or toxigenic strains which lead to wide variety of enteric and extraintestinal diseases in animals (Fairbrother et al. 2002, Asmelash, 2015).
E. coli have several types of strains that are divided into six groups of pathotypes based on the mechanism of disease cause. Enteropathogenic E. coli (EPEC), Attaching and effacing E. coli (A/EEC), Enterotoxigenic E. coli (ETEC), Enteroinvasive E. coli (EIEC), EHEC and Enteroaggregative E. coli (EAEC). E. coli strains that produce the Stx toxins have been referred to as Vero Toxin-producing E. coli (VTEC), Shiga-toxigenic E. coli (STEC) and enterohaemorrhagic E. coli (EHEC) (Karmali, 1989; Nataro and Kaper, 1998; Asmelash, 2015; Saba et al., 2015).
E. coli O157: H7 are toxigenic strains that cause life threating syndromes and resulted in an estimated 74,000 cases and 61 deaths annually in USA as a result of outbreaks arising from consumption of contaminated cattle products, especially meat and raw milk (Bedasa et al., 2018). Later, outbreaks were traced to other dairy products such as yogurt and cheese (Doyle et al., 2006; Mora et al., 2007). Escherichia coli O15 7:H7 has also been found in the intestines of healthy cattle, deer, goats, and sheep. However, cattle have been identified as a major reservoir of E. coli O157: H7 and consumption of foods of cattle origin such as beef and dairy products have been associated with some of the largest food poisoning outbreak s in which this organism was identified as the etiologic agent (Acha and Szyfress, 2001; Rey et al., 2003; Perelle et al., 2007).
Detection of E. coli O157: H7 is dependent on distinguishing the pathogenic serotypes from normal fecal flora containing commensal strains of E. coli (Battisti et al., 2006; Woynshet, 2014). Fortunately, E. coli O157: H7 has two unusual biochemical markers; delayed fermentation of D-sorbitol and lack of β-D-glucuronidase activity, which help to phenotypically separate E. coli O157: H7 isolates from nonpathogenic E. coli strains. One of these markers (delayed sorbitol fermentation) enables to develop of several selective media (Sorbitol-MacConkey) which aid in the initial recognition of suspicious colonies isolated from bloody stools (Woynshet, 2014). Detection of E. coli O157: H7 from food samples requires enrichment and isolation with selective and/or indicator media, but lacks specificity to identify STEC (Ji-Yeon et al., 2005).
Even though milk represents an important food in consumers’ nutrition as well as in the nutrition and income of producers, there is limited work so far undertaken regarding the isolation and identification of E.coli from raw cow milk in Bishoftu town. Therefore, this was done in the manner of studying the occurrence of E. coli in raw milk samples through a series of biochemical tests.
Materials and Methods
Study Area
The study was conducted to isolate and identify Escherichia coli from raw milk of bovine in selected dairy farms of central Bishoftu. The study area was selected based on the abundance of dairy farms in the area.
The study was conducted to isolate and identify Escherichia coli from raw milk of bovine in selected dairy farms of central Bishoftu. The study area was selected based on the abundance of dairy farms in the area.
Study Animal
Privately owns small holder dairy farms and state dairy farms located in Bishoftu town were involved in the study population. The study considered apparently healthy lactating dairy cows, of different breed of cattle at different age category.
Privately owns small holder dairy farms and state dairy farms located in Bishoftu town were involved in the study population. The study considered apparently healthy lactating dairy cows, of different breed of cattle at different age category.
Study Design and Sampling Strategy
A cross-sectional study was conducted from January to April 2018 to isolate and identify E. coli from raw cow milk from selected dairy farms of Bishoftu, Ethiopia. The farms were selected by using simple random sampling strategies based on data obtained from the Bishoftu town Agricultural and natural resource development office, department of livestock and fishery. A total of 50 samples of raw milk were collected from the selected dairy farms. Data regarding the different host risk factors (age, breed and body condition) were collected from apparently healthy lactating dairy cows based on the clinical examination of the animal.
A cross-sectional study was conducted from January to April 2018 to isolate and identify E. coli from raw cow milk from selected dairy farms of Bishoftu, Ethiopia. The farms were selected by using simple random sampling strategies based on data obtained from the Bishoftu town Agricultural and natural resource development office, department of livestock and fishery. A total of 50 samples of raw milk were collected from the selected dairy farms. Data regarding the different host risk factors (age, breed and body condition) were collected from apparently healthy lactating dairy cows based on the clinical examination of the animal.
Milk Sample Collection
Raw milk samples were collected from lactating of dairy cows. During sampling, the sample was collected aseptically and put in to sterile screw capped bottle and kept in an ice box containing ice packs and taken immediately to microbiology laboratory for bacteriological analysis. Then the sample was stored over night in refrigerator at 4°C and processed within 24hr of sampling. Isolation and identification of bacteria was done according to the techniques recommended by Quinn et al., 2002.
Raw milk samples were collected from lactating of dairy cows. During sampling, the sample was collected aseptically and put in to sterile screw capped bottle and kept in an ice box containing ice packs and taken immediately to microbiology laboratory for bacteriological analysis. Then the sample was stored over night in refrigerator at 4°C and processed within 24hr of sampling. Isolation and identification of bacteria was done according to the techniques recommended by Quinn et al., 2002.
Bacteriological Examination of Milk Sample
Culturing was carried out according to standard protocols ISO -16654: 2001. The milk samples were stirred in a sterile Typtone soya broth in 1:10 ratio and incubated aerobically at 37?C for 18-24 hrs. A loopfull from the incubated broth was streaked onto MacConkey agar base (Oxoid) and incubated at 37?C for 24-48 hrs. All suspected colonies that shows characterstic features of E.coli from each plate were picked up, streaked on buffered peptone water (BPW) and incubated at 37?C for 18-24 hrs for further identification.
Culturing was carried out according to standard protocols ISO -16654: 2001. The milk samples were stirred in a sterile Typtone soya broth in 1:10 ratio and incubated aerobically at 37?C for 18-24 hrs. A loopfull from the incubated broth was streaked onto MacConkey agar base (Oxoid) and incubated at 37?C for 24-48 hrs. All suspected colonies that shows characterstic features of E.coli from each plate were picked up, streaked on buffered peptone water (BPW) and incubated at 37?C for 18-24 hrs for further identification.
Isolation and Identification of Escherichia coli
For the isolation and identification of E. coli, one ml from incubated BPW was transferred to Eosin methylene blue (EMB) (Oxoid) agar and incubated at 37?C for 24 hrs. Morphologically typical colonies producing metallic sheen were taken sorbitol MacConkey agar base (Oxoid) and incubated at 37?C for 24 hr. The purified colonies were then streaked onto nutrient broth and incubated at 37?C for 18-24 hrs for further identification.
For the isolation and identification of E. coli, one ml from incubated BPW was transferred to Eosin methylene blue (EMB) (Oxoid) agar and incubated at 37?C for 24 hrs. Morphologically typical colonies producing metallic sheen were taken sorbitol MacConkey agar base (Oxoid) and incubated at 37?C for 24 hr. The purified colonies were then streaked onto nutrient broth and incubated at 37?C for 18-24 hrs for further identification.
Simultaneously another single colony with similar characteristics was picked from agar plate and stained with Gram’s stain. The isolate was examined for stain and morphological characteristics using bright-field microscopy. KOH test was then employed to confirm the Gram’s reaction (Quinn et al., 2004). Suspected colonies of E. coli (pinkish color appearance on MacConkey agar and green metallic sheen on Eosin Methylene Blue was then sub-cultured onto nutrient agar to appreciate colony characteristics and then pure colonies taken from EMB was inoculated on nutrient agar (non-selective media). The isolated colonies were subjected to series of different biochemical tests.
Biochemical Characteristics
The isolated strains were subjected to a series of different biochemical tests using the procedure of (ISO, 2003) to confirm E. coli. Catalase test, indole production test, Methyl red test, Voges-Proskauer test, and Simmon’s citrate test on tryptone broth, MR-VP medium, Simon citrate agar and Sugar fermentation were performed on all suspected isolates to confirm the E. coli.
The isolated strains were subjected to a series of different biochemical tests using the procedure of (ISO, 2003) to confirm E. coli. Catalase test, indole production test, Methyl red test, Voges-Proskauer test, and Simmon’s citrate test on tryptone broth, MR-VP medium, Simon citrate agar and Sugar fermentation were performed on all suspected isolates to confirm the E. coli.
Data Management and Statistical Analysis
Data collected from field and laboratory investigations were recorded, screened and coded using Microsoft Excel 2013 program and analyzed using STATA version 12.0. Descriptive statistics were used to figure out the proportions of E.coli isolate. The Chi-square(x2) test was applied to test the existence of association between the pathogen and the risk factors and a p-value less than 0.05 was taken as statistically significant.
Data collected from field and laboratory investigations were recorded, screened and coded using Microsoft Excel 2013 program and analyzed using STATA version 12.0. Descriptive statistics were used to figure out the proportions of E.coli isolate. The Chi-square(x2) test was applied to test the existence of association between the pathogen and the risk factors and a p-value less than 0.05 was taken as statistically significant.
Results
E.coli isolation and identification
The results of the present study revealed that out of 50 samples, 20 samples were found to be positive for E. coli. Isolates were characterized as bright pink color on MacConkey agar plates (Figure 1A) and showed blue-greenish metallic sheen on EMB agar plate (Figure 1B). Upon Gram’s staining of the isolates under 100x using light microscope, pink-colored, small rod-shaped organisms arranged in single, pairs or short-chain were identified (Figure 1C).
The results of the present study revealed that out of 50 samples, 20 samples were found to be positive for E. coli. Isolates were characterized as bright pink color on MacConkey agar plates (Figure 1A) and showed blue-greenish metallic sheen on EMB agar plate (Figure 1B). Upon Gram’s staining of the isolates under 100x using light microscope, pink-colored, small rod-shaped organisms arranged in single, pairs or short-chain were identified (Figure 1C).
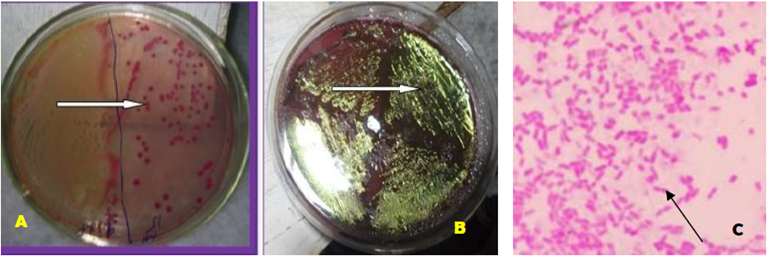
Figure 1: (A) Pink colonies of E. coli on MacConkey agar, (B). Metallic sheen on EMB by E. coli. (C). Gram-negative, pink-colored, small rod shape E. coli (arrow) (100x).
Biochemical characterization of E. coli
The biochemical characteristics of E. coli isolate showed positive for catalase, Methyl red and indole test but negative for Voges-Proskauer, urease, and citrate. In addition, reactions in TSI agar slant revealed yellow but with gas and production of hydrogen sulfide was observed. Almost all the isolates of E. coli fermented lactose, sucrose and glucose with the production of both acid and gas (Table 1).
The biochemical characteristics of E. coli isolate showed positive for catalase, Methyl red and indole test but negative for Voges-Proskauer, urease, and citrate. In addition, reactions in TSI agar slant revealed yellow but with gas and production of hydrogen sulfide was observed. Almost all the isolates of E. coli fermented lactose, sucrose and glucose with the production of both acid and gas (Table 1).
| Biochemical test | Reaction | |
| Lactose fermentation | Positive | |
| Catalase | Positive | |
| Simmon’s citrate | Negative | |
| Indole Production | Positive | |
| Nitrate Reduction | Positive | |
| Methyl Red | Positive | |
| Voges- Proskauer | Negative | |
| Urease | Negative | |
| Acid from sugar | ||
| Glucose | Positive | |
| Mannitole | Positive | |
| Lactose | Positive | |
| Salicin | Positive | |
| Sucrose | Positive | |
Table 1: Biochemical characterization of E. coli.
Association of host risk factors and E.coli isolates
The present study showed a statistically significant (P=0.048) association between E.coli and body condition of animals. In this study animals with good body conditions were at high risk of infected with E.coli as compared with animals that have poor and medium body conditions (Table-2).
The present study showed a statistically significant (P=0.048) association between E.coli and body condition of animals. In this study animals with good body conditions were at high risk of infected with E.coli as compared with animals that have poor and medium body conditions (Table-2).
| Body Condition | Total animal examined | E.coli positive (%) | X2 | P-value |
| Poor | 4 | 4(8%) | 6.092 | 0.048 |
| Moderate | 12 | 4(8%) | ||
| Good | 34 | 13(26.0%) | ||
| Total | 50 | 21(42%) |
Table 2: Association of E.coli with regards to body condition.
Out of the total animal examined in the study, the recovery rate of E.coli was 34% in young animal compared with adult animals (8%) and this shows statistically significant (P=0.014) association between E.coli and age. Similarly, out of overall examined animals, 34% of crossbreeds were positive for E.coli as compared with Holstein (8%). This result showed that there was a statistically significant association between E.coli and breed (Table 3).
| Age | Total animal examined | E.coli positive n (%) | X2 | P-Value | |
| Young | 46 | 17 (34) | 6.004 | 0.014 | |
| Adult | 4 | 4 (8) | |||
| Breed | |||||
| Cross | 46 | 17 (34) | |||
| Holstein | 4 | 4 (8) | |||
| Total | 50 | 21(42%) | |||
Table 3: Association of E.coli isolates with age and breed.
Discussion
In current study, a total of Fifty raw milk samples were collected and processed bacteriologically and biochemical tests were performed to detect Escherichia coli from raw milk. All the E. coliisolates were able to produce bright pink colored colonies on MacConkey agar, characteristic metallic sheen colonies on the EMB agar and pink colored, small rod-shaped Gram-negative bacilli on Gram’s staining. The results of catalase, MR and indole test of the E. coli isolates were positive but the V-P test was negative which are in agreement with the reports of (Zinnah et.al., 2007).
The pattern of sugar fermentation reaction by the isolated E. coli with three sugars was observed and produced acid and gas. The isolates were able to ferment glucose, lactose, and sucrose completely. Acid production was indicated by the color change from reddish to yellow and the gas production was noted by the appearance of gas bubbles in the test tubes. This result was in agreement with the findings (Asmelash, 2015; Bedassa, 2018; Zinnash, 2007; Giwida and Gohary, 2013). This result was partially in agreement with the findings of (Beutin et al., 1993 and Sandhu et al., 1996). They reported that although E. coli ferments all three basic sugars but it partially ferments sucrose and glucose. Variation of the results may be due to genetic factors and the nature of the inhabitant of the organisms.
In this study, 42% of the raw milk were contaminated with E. coli. In addition, three factors on the farm level were assessed as probable variables related to the higher frequency of samples positive for E.coli. There was statistically significant (P< 0.05) associations between body condition, age and breeds of the animals with positive isolates. This finding was comparable with the finding of Iqbal et al. 2004 (40.7%). However, it is much higher than the finding of (Biruke and Shimeles, 2015) (18.6%). This prevalence of Escherichia coli is presumably due to the fact that E. coli is the commonest environmental contaminants, which is closely associated with hygiene condition of the animals as well as the environment. It becomes pathogenic whenever the hygienic conditions of the animal or environment become poor. Moreover, the existence of high concentration of E. coli in milk also indicates the relatively poor quality of milk, related with substandard hygiene of the farm management, milk collection and processing system. The isolation of E. coli is of public health significance as this bacterium is known to cause serious gastrointestinal disorders in both young and adult humans (FAO and WHO, 2004).
Concerning the type of examined milk samples, the high prevalence of E. coli in raw milk may be attributed in Bishoftu dairy farms is since milk is mainly transported directly to the dairy plant for processing meanwhile market milk is usually collected from small farms or farmers therefore it will be liable to cross contamination by different ways as mixed fresh clean milk with mastitis milk, unclean hands of workers, unclean utensils and unhygienic water supply for washing the utensils could be the source for accelerating the bacterial contamination. This idea agreed with conditions for contamination of raw milk at different critical points due to less hygienic practices (Reta et al., 2016; Gwida and EL-Gohary, 2013).
Conclusion and Recommendations
The present study was conducted on isolation and identification of E.coli from dairy cow raw milk in Bishoftu from January to April 2018 and the prevalence of E.coli from the collected raw milk was 42%. This indicates that E.coli is one of the major problems of dairy cows in milk production that reduced the quality of milk. The distribution of this bacterial pathogen in the herd indicates the economic impact of the disease. Besides, the disease has economic importance and it also do harm the health and well being of human being. In conclusion, the results of the present study provided that microbial quality and safety of raw milk was unsatisfactory. These findings stress the need for an integrated control of E. colifrom farm production on to consumption of food of animal origin. In light of the above conclusive remarks, the following recommendations are forwarded:-
- Awearness should be created on milk handling practice, storage and milking process to Dairy farmer and milk collectors.
- The professionals should apply different methods for prevention and control of the disease.
- The professionals should inform the public about the relevance of milk pasteurization before consumption to avoid food born infection.
References
- Abebe, M., Hailelule, A., Abrha, B., Nigus, A., Birhanu, M., Adane, H., Genene, T., Getachew, G., Merga, G. and Haftay, A. (2014). Antibiogram of Escherichia coli strains isolated from food of bovine origin in selected Woredas of Tigray, Ethiopia. African Journal of Bacteriology Research, 6: 17-22.
- Abera, L. (2008). Study on Milk Production and Traditional Dairy Handling Practices in East Shoa Zone, Ethiopia [M.S. thesis], Faculty of Veterinary Medicine, Addis Ababa University. Pp:55-91
- Acha, P. and Szyfress, B. (2001). Zoonoses and communicable diseases common to man and animals, Bacteriosis and mycoses. 3rd ed. Washington, D.C: Pan American Sanitary Bureau, Pp: 12-30.
- Asmelash, T. (2015). Isolation, Identification, Antimicrobial Profile and Molecular Characterization of Enterohaemorrhagic E. coli O157: H7 Isolated from Ruminants Slaughtered at Debre Zeit Elfora Export Abattoir and Addis Ababa Abattoirs Enterprise. [M.S.c thesis], Faculty of Veterinary Medicine, Addis Ababa University.
- Battisti, A., Lovari, S., Franco, A., Diegidio, A., Tozzoli, R., Caprioli, A.and Morabito, S. (2006). Prevalence of Escherichia coli O157 in Lambs at Slaughter in Rome, Central Italy. Epidemiology of Infection, 134: 415–419.
- Bedasa, S., Daniel, S., Ashebr, A.and Tesfanesh, M. (2018): Occurrence and Antimicrobial Susceptibility Profile of Escherichia Coli O157: H7 from Food of Animal Origin in Bishoftu Town, Central Ethiopia. International Journal of Food Contamination 5:2.
- Beutin, L., Geier, D., Steinrück, H., Zimmermann, S. and Scheutz, F., (1993). Prevalence and some properties of verotoxin (Shiga-like toxin)-producing Escherichia coli in seven different species of healthy domestic animals. Journal of clinical microbiology, 31(9):2483-2488.
- Biruke, D. and Shimeles, A. (2015). Isolation and Identification of Major Bacterial Pathogen from Clinical Mastitis Cow Raw Milk in Addis Ababa, Ethiopia. Academic Journal of Animal Diseases 4(1): 44-51.
- Carbas, B., Cardoso, L., Coelho, A. (2012). Investigation on the knowledge associated with foodborne diseases in consumers of northeastern Portugal. Food Control. 30(1): 54-7.
- Doyle, M., Archer, J., Kaspar, C. and Weiss, R. (2006). Human illness caused by E. coli O157: H7 from food and non-food sources. FRI briefings.
- Fairbrother, J., Batisson, I., Girard, F., Mellata, M. and Pérès, S. (2002). Original text on E. coli. Animal Health and Production Compendium, CD-ROM CAB International, Pp:141-147
- FAO and WHO, (2004): Code of hygienic practice for milk and milk products, AC/RCP 57, Codex Alimentarius, Rome, Italy
- Gwida, MM. and EL-Gohary, FA. (2013). Zoonotic Bacterial Pathogens Isolated from Raw Milk with Special Reference to Escherichia coli and Staphylococcus aureus in Dakahlia Governorate, Egypt. Open Access Scientific Reports, 2: 705
- Iqbal, M.A.L.I., Khan, M.A., Daraz, B. and Siddique, U., (2004). Bacteriology of mastitic milk and in vitro antibiogram of the isolates. Pak. Vet.. J, 24: 161-164.
- Ji-Yeon, K., So-Hyun, K., Nam-Hoon, K., Won-Ki, B., Ji-Youn, L, Hye-Cheong, K., JunMan, K., Kyoung, N., Woo-Kyung, J., Kun-Taek, P. and Yong-Ho, P.(2005). Isolation and identification of Escherichia coli O157: H7 using different detection methods and molecular determination by multiplex PCR and RAPD. J vet sci, 6(1): 7–19.
- Karmali, M.A., (1989). Infection by verocytotoxin-producing Escherichia coli. Clinical microbiology reviews, 2(1):15-38.
- Leedom, J. (2006). Milk of non-human origin and infectious diseases in humans. Clinically Infectected Disease, 43:610-615
- LeJeune JT, Rajala-Schultz PJ (2009). Food safety: Unpasteurized milk: a continued public health threat. Clin Infect Dis 48: 93-100.
- Mora, A., León, S., Blanco, M., Blanco, J., López, C., Dahbi, G., Echeita, A., González, E. and Blanco, J. (2007). Phage types, virulence genes and PFGE profiles of shiga toxin-producing E.coli O157: H7 isolated from raw beef, soft cheese and vegetables in Lima (Peru). International Journal Food Microbiology, 114(2): 204-253.
- Nataro, J.P. and Kaper, J.B., (1998). Diarrheagenic Escherichia coli. Clinical microbiology reviews, 11(1):142-201.
- Nigatu, D., Birhanu, S., Shimelis, M., Yimer, M., and Dinaol, B. (2017). Prevalence and Antimicrobial Susceptibility Pattern of E. coli O157: H7 Isolated from Traditionally Marketed Raw Cow Milk in and around Asosa Town, Western Ethiopia. Hindawi Veterinary Medicine International Volume, Article ID 7581531, 7
- Oliver, S.P., Jayarao, B.M. and Almeida, R.A., (2005): Foodborne pathogens in milk and the dairy farm environment: food safety and public health implications. Foodbourne Pathogens & Disease, 2(2):115-129.
- Perelle, S., Dilasser, F., Grout, J., and Fach, P. (2007). Screening food raw materials for the presence of the world’s most frequent clinical cases of Shiga toxin encoding Escherichia coli I O26, O103, O111, O145 and O157. International Journal Food Microbiology, 11:284-288.
- Quinn, P., Carter, M., Markey, B. and Carter, G (2002). Clinical veterinary microbiology. Mosby International Limited: Spain, Pp. 96 -344.
- Reta, M.A., Bereda, T.W. and Alemu, A.N., (2016). Bacterial contaminations of raw cow’s milk consumed at Jigjiga City of Somali Regional State, Eastern Ethiopia. International Journal of Food Contamination, 3(1): 4.
- Rey, J., Blanco, J.E., Blanco, M., Mora, A., Dahbi, G., Alonso, J.M., Hermoso, M., Hermoso, J., Alonso, M.P., Usera, M.A. and González, E.A., (2003). Serotypes, phage types and virulence genes of Shiga-producing Escherichia coli isolated from sheep in Spain. Veterinary microbiology, 94(1): 47-56.
- Saba CKS, Eric Y and Adzitey F. (2015). Prevalence of Escherichia coli and Shiga Toxin-Producing Escherichia coli in Cattle Faeces and Raw Cow Milk Sold in the Tamale Metropolis, GHANA. Journal Dairy Veterinary Animal Research, 2(5): 37-92
- Shunda, D., Habtamu, T.and Endale, B. (2013). Assessment of bacteriological quality of raw cow milk at different critical points in Mekelle, Ethiopia, International Journal of Livestock Research, 3(4).42-48.
- Sandhu, K.S. Clark, R.C. McFadden, K. Brouwer, A. Louie, M. Wilson, J. Lior, H. Gyles, C.L. (1996). Prevalence of the eaeA gene in verotoxigenic Escherichia coli strains from dairy cattle in Southwest Ontario Epidemiol. Infect., 116:1-7
- Tchaptchet, S. and Hansen, J., (2011). The Yin and Yang of host-commensal mutualism. Gut Microbes, 2(6): 347-352.
- Virpari, P., Nayak, J., Thaker, H.and Brahmbhatt, M. (2013). Isolation of pathogenic Escherichia coli from stool samples of diarrhoeal patients with history of raw milk consumption, Veterinary World, 6(9): 659-663
- Woynshet, H. (2014). Escherichia Coli O157:H7: Prevalence and Sources of Contamination of Cattle Meat at Municipal Abattoir and Butcheries as well as its Public Health Importance in Addis Ababa, Ethiopia [M.S. thesis], Faculty of Veterinary Medicine, Addis Ababa University, Pp. 25-56.
- Yilma, Z. and Faye, B., (2006). Handling and microbial load of cow’s milk and Irgo-fermented milk collected from different shops and producers in central highlands of Ethiopia. Ethiopian Journal of Animal Production, 6(2): 67-82.
- Zinnah, M.A., Bari, M.R., Islam, M.T., Hossain, M.T., Rahman, M.T., Haque, M.H., Babu, S.A.M., Ruma, R.P. and Islam, M.A., (2007). Characterization of Escherichia coli isolated from samples of different biological and environmental sources. Bangladesh Journal of Veterinary Medicine, 5 (1&2): 25-32.
Citation: Rundasa Megersa, Mesfin Mathewos, Haben Fesseha. (2019). Isolation and Identification of Escherichia coli from Dairy Cow Raw Milk in Bishoftu Town, Central Ethiopia. Archives of Veterinary and Animal Sciences 1(1).
Copyright: © 2019 Haben Fesseha. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
