Research Article
Volume 1 Issue 2 - 2019
Cytotoxicity of New Damsin Derivatives in Breast Cancer Cells
1Centre for Analysis and Synthesis, Lund University, P.O.Box 124, 22100 Lund, Sweden
2Department of Biology, Lund University, Lund, Sweden
3Instituto de Investigaciones Químicas, Universidad Mayor de San Andrés, La Paz, Bolivia
4Instituto de Biología Molecular y Biotecnología, Universidad Mayor de San Andrés, La Paz, Bolivia
2Department of Biology, Lund University, Lund, Sweden
3Instituto de Investigaciones Químicas, Universidad Mayor de San Andrés, La Paz, Bolivia
4Instituto de Biología Molecular y Biotecnología, Universidad Mayor de San Andrés, La Paz, Bolivia
*Corresponding Author: Olov Sterner, Centre for Analysis and Synthesis, Lund University, P.O.Box 124, 22100 Lund, Sweden.
Received: August 19, 2019; Published: August 27, 2019
Abstract
As a follow-up of a previous investigation in which semisynthetic damsin derivatives were shown to possess up to 10 times higher cytotoxicity in JIMT-1 breast cancer cells compared to normal breast epithelial MCF-10A cells, a range of new derivatives were prepared and assayed toward the same cells. Damsin, a natural plant metabolite containing a??-methylene-?-lactone (or 3-methylenedihydrofuran-2(3H)-one) moiety, was modified in position 3 by Claisen-Schmidt condensations with aromatic aldehydes, mainly mono- or disubstituted benzaldehydes, without affecting the α-methylene-γ-lactone function. This lactone ring is a Michael acceptor that is known to affect biological processes such as cell proliferation, death/apoptosis, and cell migration, by interfering with nucleophilic sites in cell signalling pathways. However, although Michael acceptors are reactive, the Michael addition is reversible and it can be assumed that also other parts of the molecules will moderate the binding to and the release from any given nucleophilic site in a protein, and thereby moderate a specific biological activity. In this investigation, the cytotoxicity of 20 α-methylene-γ-lactones towards normal breast epithelial MCF-10A cells as well as breast cancer JIMT-1 cells is compared, by determining the inhibitory concentration 50 (IC50) from dose response curves. The IC50 values in the two cell lines were found to depend on the overall structure of the assayed compounds, although less in this subset of compounds compared to a previous investigation. Structure-activity relationships that may explain the observed differences in potency and selectivity are discussed.
Keywords: MCF-10A; JIMT-1; Semisynthesis; α-methylene-γ-lactones; Michael acceptors; SAR:
Introduction
Breast cancer is one of the most common forms of cancer and established as a principal cause of death for women. In 2016, more than 500 000 women in the world died from this disease, which is expected to increase over the coming years. [1] At least 20% of all breast cancer cells show overexpression of human epidermal growth factor receptor 2 (HER2) protein, and HER2 overexpression has repeatedly been identified as a factor indicating poor prognosis. [2] Other proteins also involved in breast cancers are transcription factors as well as proteins involved in the cellular signalling pathways. Although there are traditional methods for the treatment of breast cancer such as surgery, chemotherapy and radiotherapy, these are not efficient enough. Novel and effective treatments or more potent drugs with less side effects specifically targeting breast cancer are urgently needed. [3]
Secondary plant metabolites have always been an important provider of low-molecular anti-cancer drugs, and are expected to be so also in the future. [4,5,6] Our understanding of how cancer cells are formed and tumours develop and progress, as well as how treatment resistance may develop, has increased, not the least on a molecular level. This should enable us to design new and better drugs. [7] A cellular molecular pathway important for the development of chemoresistance and metastasis in many cancer cells is the NF-kB pathway, which the activation or the over-expression of is a harmful property of tumour cells. [7] Terpenoids with an a-methylene-g-lactone moiety have been shown to possess promising anti-cancer effects, interfering with biological processes such as cell signalling, proliferation, death/apoptosis, and migration. [8,9] Such terpeniods are most commonly sesquiterpens, and sesquiterpene lactones (SLs) is a large group of natural products that have been isolated from numerous genera of Asteraceae family. [10] Modern pharmacological studies have revealed their diverse biological activities, including anti-tumoral, anti-inflammatory, anti-feedant, anti-microbial and anti-protozoal. [11,12] The significant cytotoxic activity of such terpenes is linked to the α-methylene-β-lactone moiety, which via a Michael addition can alkylate nucleophilic residues (primarily free thiols of cysteines) in proteins. A target suggested for such an interaction is the protein p65 in the heterodimeric transcription factor NF-kB pathway. [13] p65 is one of the key regulator of genes involved in a broad range of biological activities such as immune responses, development, survival, proliferation, angiogenesis, invasion and metastasis. [14] A well-known SL is thapsigargin, which has undergone phase I clinical trials for breast, kidney and prostate cancer treatments.9
Electrophilic compounds are considered potentially toxic and have been rejected by the pharmaceutical industry in their search for novel drug candidates. However, 39 electrophilic drugs, mainly used in oncological therapies, have been approved by the US Food and Drug Administration. [1,16] These may display off-target toxicity, [8] but the efficiency and selectivity of an electrophilic compound will besides the reactivity of the electrophilic moiety depend on the affinity to the target through non-covalent interactions. Michael acceptors will react with nucleophiles via an addition reaction, not a substitution, in a reaction that is reversible, and will in principle bind tighter to nucleophiles that present a more favourable molecular environment.
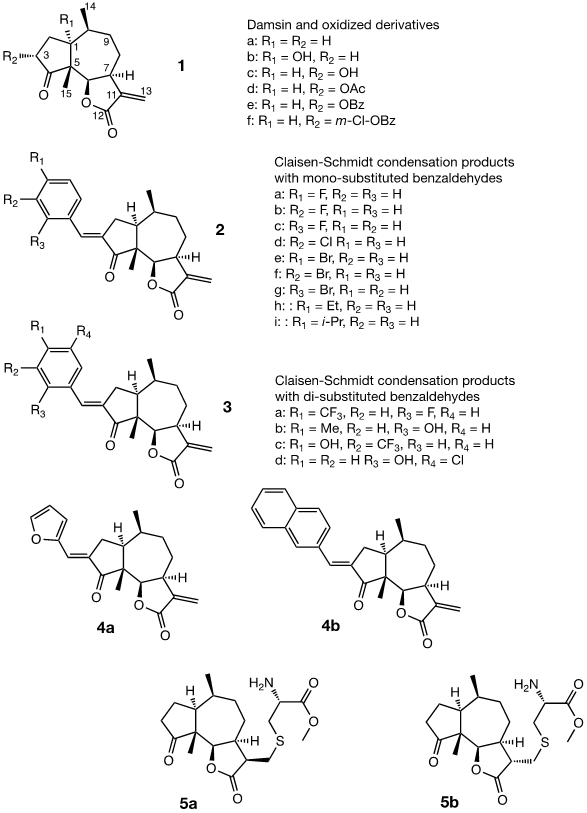
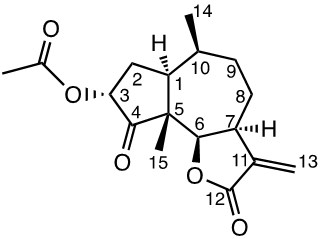
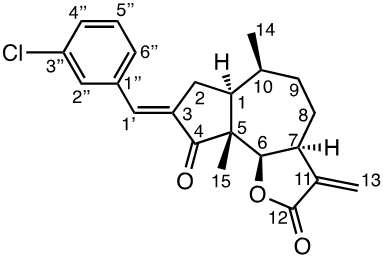
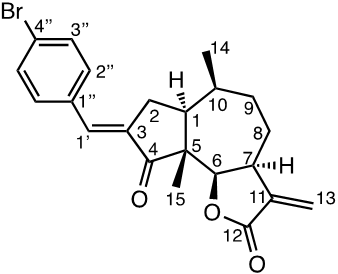
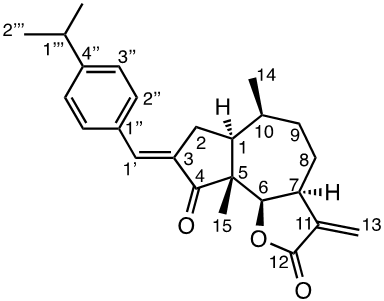
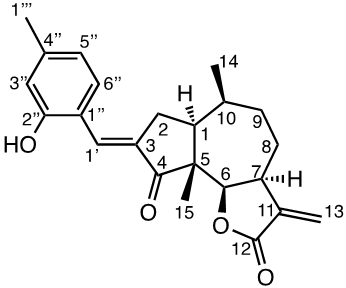
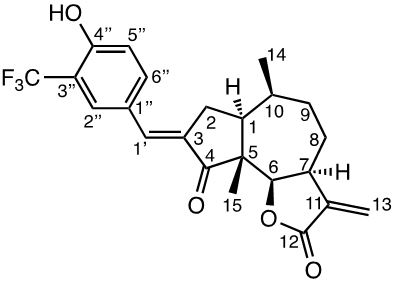
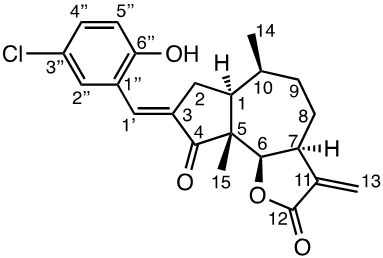
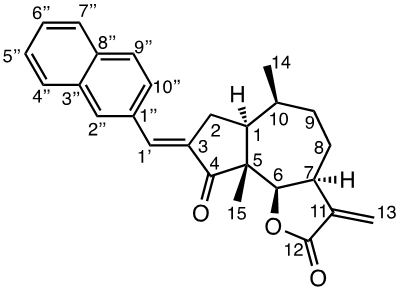
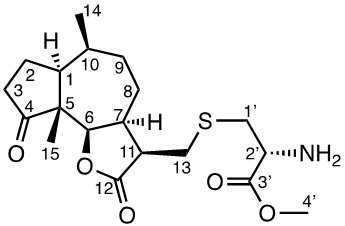
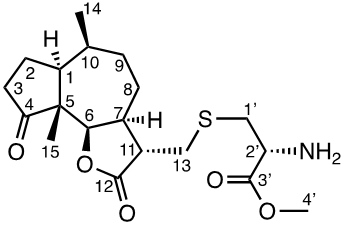
Damsin (1a) and coronopilin (1b) are pseudoguaianolide SLs, present in the plant Ambrosia arborescens. A spectrum of biological activities of damsin have been reported, e.g. an inhibitory effect on colon cancer cell lines (Caco-2 cell), as well as an influence on DNA biosynthesis and expression of NF-kB and STAT3 patways. [17,18] In the present study we compare the cytotoxicity of 20 a-methylene-g-lactones based on the natural product damsin (1a) (see Figure 1), in normal breast epithelial MCF-10A cells and breast cancer JIMT-1 cells. The compounds were prepared by Claisen-Schmidt condensations of 1a with aromatic aldehydes, as well as derivatives of 3-hydroxyldamsin (3c) prepared by oxidation of 1a. In addition, the two thioether adducts 5a and 5b in which cystein methyl ester by a Michael addition was added to C-13 in the a-methylene-g-lactone moiety, were prepared and assayed.
Experimental Procedures
General chemical procedures
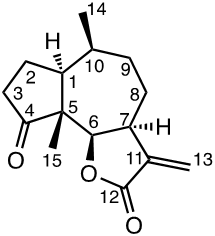
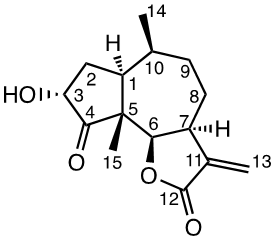
Chemicals (analytical grade) were purchased from different commercial suppliers and used without further purification. Damsin (1a) and coronopilin (1b) (Figure 1) were isolated from A. arborescens as previously described. [13] IR spectra were recorded with a Bruker Alpha FT-IR Spectrometer, and the optical rotations were measured with a Perkin-Elmer 141 polarimeter. HRMS spectra were recorded with a Waters XEVO-G2 QTOF instrument, while NMR spectra were recorded in CDCl3 using a Bruker DRX spectrometer operating at 400 MHz for 1H and at 100 MHz for 13C. Chemical shifts are given in ppm relative to the solvent signals (7.26 ppm for 1H and 77.00 ppm for 13C). All compounds described here were completely analysed by 2D NMR (COSY, HMQC, HMBC and NOESY), and as the syntheses start with the pure enantiomer damsin (1a) all compounds are given with the absolute configuration in Figures 1 and 2. Chromatography was performed with 60 Å 30-75 μm silica gel, while TLC analyses were made on Silica Gel 60 F254 (Merck) plates.
Chemicals (analytical grade) were purchased from different commercial suppliers and used without further purification. Damsin (1a) and coronopilin (1b) (Figure 1) were isolated from A. arborescens as previously described. [13] IR spectra were recorded with a Bruker Alpha FT-IR Spectrometer, and the optical rotations were measured with a Perkin-Elmer 141 polarimeter. HRMS spectra were recorded with a Waters XEVO-G2 QTOF instrument, while NMR spectra were recorded in CDCl3 using a Bruker DRX spectrometer operating at 400 MHz for 1H and at 100 MHz for 13C. Chemical shifts are given in ppm relative to the solvent signals (7.26 ppm for 1H and 77.00 ppm for 13C). All compounds described here were completely analysed by 2D NMR (COSY, HMQC, HMBC and NOESY), and as the syntheses start with the pure enantiomer damsin (1a) all compounds are given with the absolute configuration in Figures 1 and 2. Chromatography was performed with 60 Å 30-75 μm silica gel, while TLC analyses were made on Silica Gel 60 F254 (Merck) plates.
Synthetic Procedures
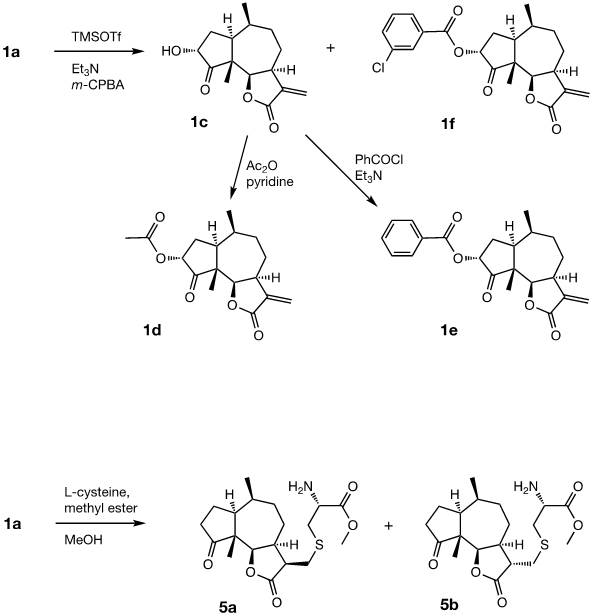
Preparation of 3α-hydroxydamsin (1c), 3α-acetoxydamsin (1d), 3α-benzoyloxydamsin (1e) and 3α-(m-chlorobenzoyloxy) damsin (1f).
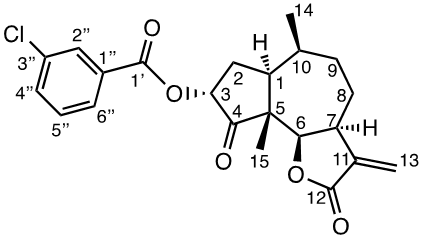
The procedures are summarized in Figure 2. To a mixture of 1a (200 mg, 0.8054 mmol) and Et3N (550 μl, 3.2216 mmol) in dry DCM (6 ml) at 0°C, TMSOTf (440 μl, 2.4162 mmol) was added slowly. The reaction was stirred at 0°C for 2h and then quenched by adding an excess of solid Na2CO3 whereafter the suspension was dried in vacuo. The dry remaining solid was extracted with hexane under stirring (3 x 20 ml x 30 min each extraction), the combined organic extracts were concentrated in vacuo to give the crude enol silane intermediate that was used in the next step without further purification. To a suspension of m-CPBA (218.4 mg, 0.8860 mmol) in n-hexane (6 ml) at -20°C, a solution of this crude enol silane intermediate in n-hexane (6.8 ml) was added dropwise. The reaction mixture was stirred for 5.5 h. The reaction was quenched with a saturated solution of Na2S2O3 (1 ml) and brine (20 ml). The aqueous mixture was extracted with DCM (3 x 20 ml), the combined organic layers were dried over Na2SO4, and concentrated in vacuo. The products were purified by silica gel flash chromatography (EtOAc: Pet.Et.40-60) to yield the products 1f (62 mg, 19%) and 1c (70 mg, 33%). To a solution of 1c (60 mg, 0, 2270 mmol) in DCM (4.6 ml) at 0°C, PhCOCl (56 µl, 0.4824 mmol) was added followed by Et3N (95 μl, 0.6834 mmol). The reaction was stirred for 23h, quenched with 5% HCl and brine (10 ml). The aqueous mixture was extracted with DCM (2 x 10 ml), the combined organic layers were dried over Na2SO4 and concentrated in vacuo. The product was purified by silica gel flash chromatography (EtOAc: n-hexane) to yield 1e (52 mg, 62%). 1d was prepared from 1c as reported previously. [19]
The procedures are summarized in Figure 2. To a mixture of 1a (200 mg, 0.8054 mmol) and Et3N (550 μl, 3.2216 mmol) in dry DCM (6 ml) at 0°C, TMSOTf (440 μl, 2.4162 mmol) was added slowly. The reaction was stirred at 0°C for 2h and then quenched by adding an excess of solid Na2CO3 whereafter the suspension was dried in vacuo. The dry remaining solid was extracted with hexane under stirring (3 x 20 ml x 30 min each extraction), the combined organic extracts were concentrated in vacuo to give the crude enol silane intermediate that was used in the next step without further purification. To a suspension of m-CPBA (218.4 mg, 0.8860 mmol) in n-hexane (6 ml) at -20°C, a solution of this crude enol silane intermediate in n-hexane (6.8 ml) was added dropwise. The reaction mixture was stirred for 5.5 h. The reaction was quenched with a saturated solution of Na2S2O3 (1 ml) and brine (20 ml). The aqueous mixture was extracted with DCM (3 x 20 ml), the combined organic layers were dried over Na2SO4, and concentrated in vacuo. The products were purified by silica gel flash chromatography (EtOAc: Pet.Et.40-60) to yield the products 1f (62 mg, 19%) and 1c (70 mg, 33%). To a solution of 1c (60 mg, 0, 2270 mmol) in DCM (4.6 ml) at 0°C, PhCOCl (56 µl, 0.4824 mmol) was added followed by Et3N (95 μl, 0.6834 mmol). The reaction was stirred for 23h, quenched with 5% HCl and brine (10 ml). The aqueous mixture was extracted with DCM (2 x 10 ml), the combined organic layers were dried over Na2SO4 and concentrated in vacuo. The product was purified by silica gel flash chromatography (EtOAc: n-hexane) to yield 1e (52 mg, 62%). 1d was prepared from 1c as reported previously. [19]
General procedure for Claisen-Schmidt/aldol condensations under acidic conditions (2e-2i and 4b)
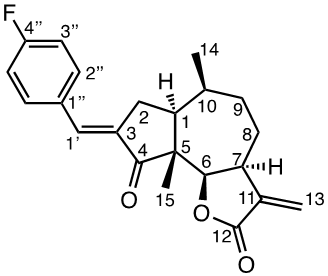
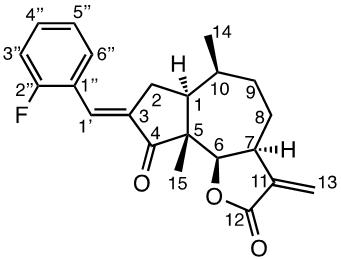
A mixture of 1a (1 eq., 0.4027 mmol), aldehyde (1.3 eq., 0.5235 mmol), and p-TsOH (1.5 eq., 0.6040 mmol) in benzene (10 ml) was prepared in a sealed tube, and stirred at 80°C for 19 to 66 h until the reaction was completed (monitored by TLC). The reaction was quenched by adding 2 ml of 5% NaHCO3 followed by 15 ml brine, and the mixture was extracted with DCM (3 x 15 ml). The combined organic layers were dried with MgSO4 and concentrated in vacuo. The crude product was purified by silica gel chromatography (EtOAc:petroleum ether 1:1) to produce the condensation products 2e (14.1 mg, 8%), 2f (58.3 mg, 35%), 2g (84.5 mg, 50%), 2h (45 mg, 30%), 2i (44.6 mg, 29%) and 4b (17.1 mg, 11%).
A mixture of 1a (1 eq., 0.4027 mmol), aldehyde (1.3 eq., 0.5235 mmol), and p-TsOH (1.5 eq., 0.6040 mmol) in benzene (10 ml) was prepared in a sealed tube, and stirred at 80°C for 19 to 66 h until the reaction was completed (monitored by TLC). The reaction was quenched by adding 2 ml of 5% NaHCO3 followed by 15 ml brine, and the mixture was extracted with DCM (3 x 15 ml). The combined organic layers were dried with MgSO4 and concentrated in vacuo. The crude product was purified by silica gel chromatography (EtOAc:petroleum ether 1:1) to produce the condensation products 2e (14.1 mg, 8%), 2f (58.3 mg, 35%), 2g (84.5 mg, 50%), 2h (45 mg, 30%), 2i (44.6 mg, 29%) and 4b (17.1 mg, 11%).
General procedure for Claisen-Schmidt/aldol condensations under basic conditions (2a-2d, 3a-3d and 4a)
A mixture of aldehyde (3 eq., 1.2081 mmol) and 1a (1 eq., 0.4027 mmol) in 50% PhMe: H2O 1:1 (3 ml) was cooled in an ice-bath, and TBAH (1.2 eq., 0.4832 mmol, 1.5 M in H2O) was added dropwise. The ice-bath was removed after 10 min and the reaction was left to reach room temperature. The reaction was monitored by TLC, and after 3 to 24h, the reaction was quenched by addition of 2 ml of 5% HCl and stirred for 15 min at r.t. The reaction mixture was diluted with 15 ml brine, and extracted with DCM (3 x 10 ml), and the combined organic layers were dried with MgSO4 and concentrated in vacuo. The crude condensation product was purified by silica gel chromatography (EtOAc:petroleum ether 1:1) to produce the condensation products 2a (22.4 mg, 39%), 2b (84.5 mg, 59%), 2c (101.5 mg, 71%), 2d (62.2 mg, 41%), 3a (100 mg, 59%), 3b (44.6 mg, 27%), 3c (61.2 mg, 32%), 3d (247 mg, 94%) and 4b (96.7 mg, 73%).
A mixture of aldehyde (3 eq., 1.2081 mmol) and 1a (1 eq., 0.4027 mmol) in 50% PhMe: H2O 1:1 (3 ml) was cooled in an ice-bath, and TBAH (1.2 eq., 0.4832 mmol, 1.5 M in H2O) was added dropwise. The ice-bath was removed after 10 min and the reaction was left to reach room temperature. The reaction was monitored by TLC, and after 3 to 24h, the reaction was quenched by addition of 2 ml of 5% HCl and stirred for 15 min at r.t. The reaction mixture was diluted with 15 ml brine, and extracted with DCM (3 x 10 ml), and the combined organic layers were dried with MgSO4 and concentrated in vacuo. The crude condensation product was purified by silica gel chromatography (EtOAc:petroleum ether 1:1) to produce the condensation products 2a (22.4 mg, 39%), 2b (84.5 mg, 59%), 2c (101.5 mg, 71%), 2d (62.2 mg, 41%), 3a (100 mg, 59%), 3b (44.6 mg, 27%), 3c (61.2 mg, 32%), 3d (247 mg, 94%) and 4b (96.7 mg, 73%).
General procedure for the MOM-protection of the hydroxybenzaldehydes.
To a solution of the hydroxybenzaldehydes corresponding to derivatives 3b-3d (2.3333 mmol) in 6 ml dry DCM was added EtN(i-Pr)2 (2 ml, 11.6665 mmol), followed by MOMBr (400 μl, 4.8999 mmol) dropwise under N2 at 0°C, whereafter the mixture was stirred at room temperature for 6 h. The reaction was quenched with 10 ml of a saturated solution of NaHCO3 by stirring for 15 min at r.t., and the mixture was extracted with DCM (2 x 15 ml). The combined organic layers were dried with MgSO4 and concentrated in vacuo. The product was purified by chromatography on silica gel (n-heptane:DCM 1:1) to yield the MOM-protected hydroxybenzaldehydes corresponding to derivatives 3b-3d.
To a solution of the hydroxybenzaldehydes corresponding to derivatives 3b-3d (2.3333 mmol) in 6 ml dry DCM was added EtN(i-Pr)2 (2 ml, 11.6665 mmol), followed by MOMBr (400 μl, 4.8999 mmol) dropwise under N2 at 0°C, whereafter the mixture was stirred at room temperature for 6 h. The reaction was quenched with 10 ml of a saturated solution of NaHCO3 by stirring for 15 min at r.t., and the mixture was extracted with DCM (2 x 15 ml). The combined organic layers were dried with MgSO4 and concentrated in vacuo. The product was purified by chromatography on silica gel (n-heptane:DCM 1:1) to yield the MOM-protected hydroxybenzaldehydes corresponding to derivatives 3b-3d.
General procedure for the deprotection of the Claisen-Schmidt adducts with MOM-protected hydroxyl groups.
To a solution of a MOM protected Claisen-Schmidt adduct, formed either by procedure 1b or 1c, (50 mg, 0.1261 mmol) in MeOH (2.6 ml), HCl conc. (22.5 μl, 0.2695 mmol, 37%) was added slowly. The reaction mixture was heated to 40°C for 5 h and then cooled to room temperature, whereafter the organic solvent was evaporated in vacuo. The residue was dissolved in DCM (15 ml), washed with brine (10 ml), dried with MgSO4, and concentrated in vacuo. The product was purified by chromatography on silica gel (EtOAc:petroleum ether 1:1) to yield the products 3b-3d that were assayed.
To a solution of a MOM protected Claisen-Schmidt adduct, formed either by procedure 1b or 1c, (50 mg, 0.1261 mmol) in MeOH (2.6 ml), HCl conc. (22.5 μl, 0.2695 mmol, 37%) was added slowly. The reaction mixture was heated to 40°C for 5 h and then cooled to room temperature, whereafter the organic solvent was evaporated in vacuo. The residue was dissolved in DCM (15 ml), washed with brine (10 ml), dried with MgSO4, and concentrated in vacuo. The product was purified by chromatography on silica gel (EtOAc:petroleum ether 1:1) to yield the products 3b-3d that were assayed.
Procedure for the preparation of the thioether adducts 5a and 5b from 1a
To a mixture of 1a (1 eq., 1.2081 mmol), L-cysteine methyl ester hydrochloride (1 eq., 1.2081 mmol) in methanol (6 ml) was added TEA (100 μl), the mixture of reaction was stirred at room temperature for 20 h until the reaction was completed (analysed by TLC). The reaction mixture was concentrated in vacuo, and the product was purified by silica gel chromatography (MeOH;DCM;TEA) to yield 45% of adduct 5a and 22.4% of adduct 5b.
To a mixture of 1a (1 eq., 1.2081 mmol), L-cysteine methyl ester hydrochloride (1 eq., 1.2081 mmol) in methanol (6 ml) was added TEA (100 μl), the mixture of reaction was stirred at room temperature for 20 h until the reaction was completed (analysed by TLC). The reaction mixture was concentrated in vacuo, and the product was purified by silica gel chromatography (MeOH;DCM;TEA) to yield 45% of adduct 5a and 22.4% of adduct 5b.
Biological assays
Cell lines
The normal-like epithelial MCF–10A cell line was purchased from American Type Culture Collection (Manassas, VA, USA) (CRL-10781) and was used as a representative for normal breast epithelial cells. The cell line retains many normal traits, including lack of tumorigenicity in nude mice, anchorage-dependent growth, and dependence on growth factors and hormones for proliferation and survival. [18,19] The MCF-10A cells were cultured in RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum (FBS), non-essential amino acids (1 mM), insulin (10 µg/ml), epidermal growth factor (20 ng/ml), cholera toxin (50 ng/ml), hydrocortisol (250 ng/ml), penicillin (100 U/ml), and streptomycin (100 µg/ml).
The normal-like epithelial MCF–10A cell line was purchased from American Type Culture Collection (Manassas, VA, USA) (CRL-10781) and was used as a representative for normal breast epithelial cells. The cell line retains many normal traits, including lack of tumorigenicity in nude mice, anchorage-dependent growth, and dependence on growth factors and hormones for proliferation and survival. [18,19] The MCF-10A cells were cultured in RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum (FBS), non-essential amino acids (1 mM), insulin (10 µg/ml), epidermal growth factor (20 ng/ml), cholera toxin (50 ng/ml), hydrocortisol (250 ng/ml), penicillin (100 U/ml), and streptomycin (100 µg/ml).
The JIMT–1 human breast carcinoma cell line (ACC589) was purchased from the German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany). It carries an amplified HER2 oncogene and is insensitive to HER2 inhibiting drugs and belongs to the HER2 sub-type of breast cancer. [20,21] The JIMT-1 cells were routinely cultured in DMEM/Ham’s F-12 medium supplemented with 10% FBS, non-essential amino acids (1 mM), insulin (10 µg/ml), penicillin (100 U/ml) and streptomycin (100 µg/ml). Both cell lines were kept at 37°C in a humidified incubator with 5% CO2 in air.
Compound solutions
The sample compounds were dissolved in DMSO to 100 mM stock solutions that were kept at -20°C. Working solutions were diluted in PBS and all had a DMSO concentration of 0.2%. In the assay, the cells were exposed to PBS with 0.02% DMSO, or with the respective compounds at 0.1, 0.25, 0.5, 1, 2.5, 5, 10, and 20 µM concentrations.
The sample compounds were dissolved in DMSO to 100 mM stock solutions that were kept at -20°C. Working solutions were diluted in PBS and all had a DMSO concentration of 0.2%. In the assay, the cells were exposed to PBS with 0.02% DMSO, or with the respective compounds at 0.1, 0.25, 0.5, 1, 2.5, 5, 10, and 20 µM concentrations.
Dose response assay
The dose response to treatment with the compounds was evaluated using an MTT assay, which is based on reduction of MTT in the mitochondria of live cells. The amount of formazan produced is proportional to the number of living cells. [21,22] For the assay, cells were trypsinized and counted in a hemocytometer. Aliquots of 180 µl cell suspension containing 3000 cells (MCF-10A) or 5000 cells (JIMT-1) were seeded in 96-well plates. Compounds were added 24 h later to the final concentrations described above. At 72 h of drug treatment, 20 µl of MTT solution (5 mg/ml in PBS) was added to each well and the 96-well plates were returned to the CO2 incubator for 1 h. The medium was then removed and the blue formazan crystals were dissolved by adding of 100 µl of 100% DMSO per well. The plates were swirled gently at room temperature for 10 minutes to dissolve the crystals. Absorbance was monitored at 540 nm in a Multiskan™ FC Microplate Photometer (Thermo Fisher Scientific, Lund, Sweden) using the software SkanIt 3.1. For each compound, three dose response experiments were performed with six replicates for each one of them. GraphPad Prism version 6.01 for Windows (GraphPad Software, La Jolla, CA, USA), was used for drawing dose response curves and calculating the IC50 values, i.e. the dose giving 50% reduction in cell number.
The dose response to treatment with the compounds was evaluated using an MTT assay, which is based on reduction of MTT in the mitochondria of live cells. The amount of formazan produced is proportional to the number of living cells. [21,22] For the assay, cells were trypsinized and counted in a hemocytometer. Aliquots of 180 µl cell suspension containing 3000 cells (MCF-10A) or 5000 cells (JIMT-1) were seeded in 96-well plates. Compounds were added 24 h later to the final concentrations described above. At 72 h of drug treatment, 20 µl of MTT solution (5 mg/ml in PBS) was added to each well and the 96-well plates were returned to the CO2 incubator for 1 h. The medium was then removed and the blue formazan crystals were dissolved by adding of 100 µl of 100% DMSO per well. The plates were swirled gently at room temperature for 10 minutes to dissolve the crystals. Absorbance was monitored at 540 nm in a Multiskan™ FC Microplate Photometer (Thermo Fisher Scientific, Lund, Sweden) using the software SkanIt 3.1. For each compound, three dose response experiments were performed with six replicates for each one of them. GraphPad Prism version 6.01 for Windows (GraphPad Software, La Jolla, CA, USA), was used for drawing dose response curves and calculating the IC50 values, i.e. the dose giving 50% reduction in cell number.
Determination of the cellular glutathione content
The cellular concentration of glutathione was determined using the GSH-Glo™ Glutathione Assay (Promega, Madison, WI, USA). The instructions of the manufacturer were followed. In short, MCF-10A cells and JIMT-1 cells were seeded at different densities (MCF-10A: 5000, 10000, and 20000. JIMT-1: 10000, 20000, and 40000) in 96-well plates in 150 ml medium and incubated for 24 hours to allow attachment of the cells. Then the glutathione assay was performed on the attached cell and in the same plate, the standard curve was generated. The glutathione content of cells seeded at different densities was obtained from the standard curve according to the instructions of the manufacturer. This data was used to calculate the intracellular concentration using cell volumes. [23] MCF-10A cells were found to have a mean volume of 998 mm3 and MCF-7 breast cancer cells a mean volume of 10124 mm3. Since we have not found any data of the volume of JIMT-1 cells but have cultured both JIMT-1 and MCF-7 cells and compared their size in the phase contrast microscope, we assume they have a similar volume. Thus, using the data from the GSH-Glo™ Glutathione Assay together with cell volume data, we have determined the glutathione concentration to be 588 ± 173 mM and 47 ± 18 mM in MCF-10A and JIMT-1 cells, respectively.
The cellular concentration of glutathione was determined using the GSH-Glo™ Glutathione Assay (Promega, Madison, WI, USA). The instructions of the manufacturer were followed. In short, MCF-10A cells and JIMT-1 cells were seeded at different densities (MCF-10A: 5000, 10000, and 20000. JIMT-1: 10000, 20000, and 40000) in 96-well plates in 150 ml medium and incubated for 24 hours to allow attachment of the cells. Then the glutathione assay was performed on the attached cell and in the same plate, the standard curve was generated. The glutathione content of cells seeded at different densities was obtained from the standard curve according to the instructions of the manufacturer. This data was used to calculate the intracellular concentration using cell volumes. [23] MCF-10A cells were found to have a mean volume of 998 mm3 and MCF-7 breast cancer cells a mean volume of 10124 mm3. Since we have not found any data of the volume of JIMT-1 cells but have cultured both JIMT-1 and MCF-7 cells and compared their size in the phase contrast microscope, we assume they have a similar volume. Thus, using the data from the GSH-Glo™ Glutathione Assay together with cell volume data, we have determined the glutathione concentration to be 588 ± 173 mM and 47 ± 18 mM in MCF-10A and JIMT-1 cells, respectively.
Results and Discussion
The two natural products damsin (1a) and coronopilin (1b) were isolated from the plant A. arborescens as previously described, [20] and 1a was used as the starting material for semisynthesis of new derivatives retaining the α-methylene-γ-lactone moiety intact. The present study is based on a previous study [21] in which Claisen-Schmidt condensation products of 1a with aromatic aldehydes forming (E)-3-benzylidendamsin derivatives were found to be more potent in the JIMT-1 cancer cells compared to the MCF-10A normal-like cells. The selectivity for the JIMT-1 cancer cells was measured as the ratio of the IC50 values for MCF-10A and JIMT-1 cells, see Table 1). Our focus in this study was therefore to prepare new condensation products between 1a and aromatic aldehydes. In general, the condensations proceeded well, and worked both under acidic and basic conditions (see Experimental procedures for more information). As noted during the previous study, it was not possible to use hydoxybenzaldehydes directly, the hydroxyl groups had to be protected as MOMO substituents prior to the condensation with 1, to eventually form compounds 3b – 3d after subsequent deprotection (as described in the Experimental procedures). As observed previously, the condensations with between 1a and aromatic aldehydes were highly stereoselective, only the E-alkenes were obtained. [21] This additional double bond, although it forms a second Michael acceptor fuctionality that theoretically could influence the biological activity, was previously shown to have no significant effect on either the potency or the selectivity. [22] In order to demonstrate the necessity of a a-methylene-g-lactone moiety for cytotoxicity of the compounds, we prepared the adducts 5a and 5b in which cysteine methyl ester was added to the the α-methylene-β-lactone moiety of damsin. This reaction proceede smoothly in methanol at room temperature, and the product could be purified by chromatography.
| Compound | MCF-10A (µM) | JIMT-1 (µM) | Ratio MCF-10A:JIMT-1 |
| 1a (damsin) | 8.1 ± 0.42 | 3.3 ± 0.62 | 2.5 |
| 1b (coronopolin) | 15.3 ± 0.9 | 5.6 ± 0.8 | 2.7 |
| 1c | 12.8 ± 1.12 | 6.3 ± 0.22 | 2.0 |
| 1d | > 201 | > 201 | na |
| 1e | > 201 | > 201 | na |
| 1f | > 201 | > 201 | na |
| 2a | 4.4 ± 1.62 | 2.3 ± 0.43 | 1.9 |
| 2b | 2.5 ± 0.32 | 1.1 ± 0.13 | 2.3 |
| 2c | 2.2 ± 0.42 | 1.1 ± 0.33 | 2.0 |
| 2d | 3.8 ± 0.62 | 1.4 ± 0.32 | 2.6 |
| 2e | 6.2 ± 1.33 | 1.8 ± 0.73 | 3.5 |
| 2f | 6.1 ± 0.43 | 1.8 ± 0.93 | 3.4 |
| 2g | 2.0 ± 0.43 | 1.2 ± 0.33 | 1.6 |
| 2h | 2.5 ± 0.42 | 1.9 ± 0.82 | 1.3 |
| 2i | 6.2 ± 1.42 | 3.8 ± 1.02 | 1.6 |
| 3a | 2.3 ± 0.92 | 1.2 ± 0.13 | 2.0 |
| 3b | 4.7 ± 0.72 | 1.9 ± 0.13 | 2.4 |
| 3c | 5.4 ± 1.43 | 1.8 ± 0.23 | 3.0 |
| 3d | 2.1 ± 0.72 | 0.7 ± 0.12 | 2.6 |
| 4a | 7.7 ± 2.82 | 2.3 ± 0.92 | 3.3 |
| 4b | 3.2 ± 1.63 | 2.1 ± 1.03 | 1.5 |
| 5a | 16.0 ± 3.53 | 8.8 ± 1.73 | 1.7 |
| 5b | > 20 | 15.9 ± 1.43 | na |
Table 1: The cytotoxicity of compounds 2 – 8, given as IC50 values (μM) calculated from experiments with six concentrations up to 20 μM. Standard deviations were obtained from 1two dose-response curves, 2three dose-response curves, or 3four dose-response curves.
The structures of all compounds prepared in this investigation was carefully determined by 1- and 2D NMR experiments (including COSY, NOESY, HMQC and HMBC experiments), in combination with the IR and HRMS data reported in the Supporting Information. The configuration of the C-3/C-1' double bond was determined by NOESY NMR experiments. [22] The chemical shifts for all proton and carbon signals observed in CDCl3 are presented in Tables 2 and 3. In all cases, the NMR couplings observed between protons are those that we expected from our previous experience from this type of compounds, and are for reasons of space not reported here but found in the Supporting Information. The comparison of the proton and carbon shifts in Tables 2 and 3 gives some interesting information. The NMR chemical shifts, and thereby the properties including the reactivity of the compounds are influenced by the electronic properties of the chemical bonds close to a specific nucleus or involved in a reactive functionality. Assuming that the a-methylene-g-lactone moiety is critical for the cytotoxicity of the compounds assayed, it is interesting to compare the chemical shifts of 6-H, 7-H, 13-H2, C-6, C-7, C-11, C-12 and C-13 of the assayed compounds, in search for any indication that the substituent at C-3 affect the electronic properties of this lactone ring. 6-H are all close to 4.7 ppm, except in 5a and 5b which no longer possess a α-methylene-β-lactone moiety. 7-H are all close to 3.3 ppm, 13-Ha are close to 5.6 ppm, while 13-Hb are close to 6.3 ppm. The C-6 values are close to 82 ppm, C-7 close to 45 ppm, C-11 close 140 ppm, C-12 close to 170 ppm, and C-13 close to 121 ppm. So, the electronic properties of the α-methylene-β-lactone moiety remains essentially intact, and appears to be essentially independent of the C-3 substituent. The conclusion is therefore that the differences in cytotoxic activities shown in Table 1 and discussed below, depend on the molecular interaction that the C-3 substituent can provide with a protein at the site where a Michael addition can take place.
| 1-H | 2-H2 | 6-H | 7-H | 8-H2 | 9-H2 | 10-H | 13-H2 | 14-H3 | 15-H3 | 1’-H | 2’’-H | 3’’-H | 4’’-H | 5’’-H | 6’’-H | |
| 1a | 2.01 | 1.79/ 2.00 | 4.44 | 3.24 | 1.79/ 1.85 | 1.81/ 1.81 | 2.15 | 5.46/ 6.13 | 1.01 | 1.00 | - | - | - | - | - | - |
| 1b | 2.47 | 2.47/ 2.47 | 4.92 | 3.34 | 1.71/ 2.08 | 1.60/ 2.40 | 2.19 | 5.58/ 6.26 | 1.20 | 1.15 | - | - | - | - | - | - |
| 1c | 2.27 | 1.81/ 2.34 | 4.69 | 3.33 | 1.91/ 1.94 | 1.68/ 1.86 | 2.15 | 5.54/ 6.25 | 1.07 | 1.09 | - | - | - | - | - | - |
| 1d | 2.35 | 1.81/ 2.48 | 4.70 | 3.31 | 1.86/ 1.99 | 1.70/ 1.87 | 2.14 | 5.55/ 6.26 | 1.07 | 1.12 | - | - | - | - | - | - |
| 1e | 2.49 | 1.98/ 2.59 | 4.79 | 3.34 | 1.86/ 2.10 | 1.73/ 1.87 | 2.16 | 5.57/ 6.28 | 1.10 | 1.18 | - | 8.00 | 7.42 | 7.56 | 7.42 | 8.00 |
| 1f | 2.46 | 1.96/ 2.59 | 4.78 | 3.35 | 1.88/ 1.99 | 1.74/ 1.91 | 2.17 | 5.56/ 6.27 | 1.10 | 1.17 | - | 7.65 | - | 7.53 | 7.36 | 7.88 |
| 2a | 2.07 | 2.86/ 2.96 | 4.65 | 3.29 | 1.78/ 2.08 | 1.72/ 1.87 | 2.28 | 5.58/ 6.29 | 1.17 | 1.16 | 7.40 | 7.11 | 7.54 | - | 7.54 | 7.11 |
| 2b | 2.10 | 2.85/ 2.95 | 4.64 | 3.30 | 1.81/ 2.07 | 1.72/ 1.90 | 2.28 | 5.57/ 6.28 | 1.18 | 1.15 | 7.37 | 7.30 | - | 7.06 | 7.39 | 7.23 |
| 2c | 2.06 | 2.76/ 2.94 | 4.64 | 3.29 | 1.82/ 2.07 | 1.73/ 1.86 | 2.26 | 5.57/ 6.27 | 1.17 | 1.16 | 7.62 | - | 7.09 | 7.35 | 7.18 | 7.53 |
| 2d | 2.08 | 2.84/ 2.95 | 4.64 | 3.30 | 1.82/ 2.06 | 1.73/ 1.91 | 2.29 | 5.57/ 6.28 | 1.18 | 1.15 | 7.33 | 7.50 | - | 7.35 | 7.34 | 7.40 |
| 2e | 2.08 | 2.81/ 2.93 | 4.65 | 3.29 | 1.83/ 2.09 | 1.75/ 1.90 | 2.22 | 5.56/ 6.28 | 1.18 | 1.16 | 7.36 | 7.40 | 7.55 | - | 7.55 | 7.40 |
| 2f | 2.08 | 2.83/ 2.94 | 4.63 | 3.29 | 1.83/ 2.07 | 1.74/ 1.89 | 2.29 | 5.57/ 6.27 | 1.17 | 1.14 | 7.31 | 7.64 | - | 7.48 | 7.28 | 7.44 |
| 2g | 2.04 | 2.69/ 2.90 | 4.61 | 3.29 | 1.81/ 2.06 | 1.72/ 1.85 | 2.21 | 5.56/ 6.26 | 1.14 | 1.16 | 7.67 | - | 7.61 | 7.19 | 7.50 | 7.25 |
| 2h | 2.07 | 2.79/ 2.94 | 4.65 | 3.28 | 1.81/ 2.08 | 1.74/ 1.88 | 2.28 | 5.57/ 6.28 | 1.18 | 1.16 | 7.42 | 7.47 | 7.25 | - | 7.25 | 7.47 |
| 2i | 2.07 | 2.87/ 2.96 | 4.65 | 3.28 | 1.81/ 2.09 | 1.73/ 1.88 | 2.28 | 5.57/ 6.28 | 1.18 | 1.16 | 7.43 | 7.49 | 7.28 | - | 7.28 | 7.49 |
| 3a | 2.09 | 2.75/ 2.95 | 4.64 | 3.31 | 1.85/ 2.07 | 1.75/ 1.89 | 2.26 | 5.57/ 6.28 | 1.16 | 1.16 | 7.55 | - | 7.36 | - | 7.45 | 7.65 |
| 3b | 2.06 | 2.80/ 2.93 | 4.65 | 3.28 | 1.78/ 2.07 | 1.74/ 1.87 | 2.26 | 5.58/ 6.30 | 1.17 | 1.18 | 8.0 | - | 6.8 | - | 6.73 | 7.36 |
| 3c | 2.06 | 2.80/ 2.91 | 4.63 | 3.28 | 1.84/ 2.07 | 1.73/ 1.89 | 2.27 | 5.57/ 6.27 | 1.16 | 1.13 | 7.34 | 7.70 | - | - | 6.96 | 7.52 |
| 3d | 2.03 | 2.74/ 2.88 | 4.60 | 3.26 | 1.79/ 2.04 | 1.69/ 1.84 | 2.25 | 5.56/ 6.25 | 1.14 | 1.12 | 7.71 | - | 6.77 | 7.12 | - | 7.34 |
| 4a | 2.05 | 2.82/ 2.94 | 4.59 | 3.26 | 1.78/ 2.01 | 1.68/ 1.82 | 2.23 | 5.51/ 6.20 | 1.10 | 1.06 | 7.12 | - | 7,53 | 6,47 | 6,63 | - |
| 4b | 2.10 | 2.97/ 3.08 | 4.67 | 3.29 | 1.82/ 2.11 | 1.76/ 1.91 | 2.32 | 5.58/ 6.30 | 1.22 | 1.20 | 7.60 | - | - | - | - | - |
| 5a | 2.08 | 1.81/ 2.03 | 4.45 | 2.59 | 1.52/ 1.52 | 1.60/ 1.85 | 2.21 | 2.59/ 3.00 | 1.06 | 1.11 | - | - | - | - | - | - |
| 5b | 2.05 | 1.83/ 1.98 | 4.41 | 2.79 | 1.78/ 1,87 | 1.66/ 1.74 | 2.18 | 2.96/ 2.87 | 1.07 | 1.08 | - | - | - | - | - | - |
In 1d, the acetyl signal appears at 2.08 ppm.
In 4b, the naphthyl signals appear at 7.51, 7.53, 7.83, 7.86, 7.89, 7.66 and 8.01 ppm.
In 5a, the cysteine signals appear at 2.78, 3.65 and 3.70 ppm, while the 11-H signal appears at 2.93 ppm.
In 5b, the cysteine signals appear at 2.98, 3.74 and 3.73 ppm, while the 11-H signal appears at 2.64 ppm.
Table 2: 1H NMR chemical shifts (in ppm) for the assayed compounds 1a-f, 2a-i, 3a-d, 4a-b and 5a-b determined at 400 MHz in CDCl3.
In 4b, the naphthyl signals appear at 7.51, 7.53, 7.83, 7.86, 7.89, 7.66 and 8.01 ppm.
In 5a, the cysteine signals appear at 2.78, 3.65 and 3.70 ppm, while the 11-H signal appears at 2.93 ppm.
In 5b, the cysteine signals appear at 2.98, 3.74 and 3.73 ppm, while the 11-H signal appears at 2.64 ppm.
Table 2: 1H NMR chemical shifts (in ppm) for the assayed compounds 1a-f, 2a-i, 3a-d, 4a-b and 5a-b determined at 400 MHz in CDCl3.
| C-1 | C-2 | C-3 | C-4 | C-5 | C-6 | C-7 | C-8 | C-9 | C-10 | C-11 | C-12 | C-13 | C-14 | C-15 | C-1’ | C-1’’ | C-2’’ | C-3’’ | C-4’’ | C-5’’ | C-6’’ | |
| 1a | 45.8 | 25.5 | 36.0 | 218.9 | 54.7 | 81.6 | 44.2 | 23.8 | 33.2 | 34.1 | 139.5 | 170.1 | 120.6 | 15.7 | 13.6 | - | - | - | - | - | - | - |
| 1b | 84.2 | 32.3 | 31.0 | 216.4 | 57.9 | 78.4 | 43.9 | 26.8 | 29.4 | 42.1 | 140.0 | 169.4 | 120.7 | 16.5 | 13.9 | - | - | - | - | - | - | - |
| 1c | 42.3 | 31.2 | 70.1 | 216.9 | 54.8 | 81.7 | 43.7 | 25.4 | 33.4 | 33.9 | 139.6 | 170.1 | 121.1 | 14.9 | 13.7 | - | - | - | - | - | - | - |
| 1da | 42.6 | 30.1 | 71.5 | 211.8 | 55.2 | 81.3 | 44.0 | 25.9 | 34.0 | 33.7 | 139.7 | 170.0 | 121.3 | 15.0 | 14.1 | - | - | - | - | - | - | - |
| 1e | 42.8 | 30.3 | 72.2 | 211.8 | 55.4 | 81.5 | 44.1 | 26.0 | 34.0 | 33.8 | 139.8 | 170.1 | 121.4 | 15.2 | 14.3 | 165.7 | 129.3 | 129.9 | 128.5 | 133.5 | 128.5 | 129.9 |
| 1f | 42.7 | 30.0 | 72.3 | 211.3 | 55.3 | 81.3 | 44.0 | 25.9 | 33.9 | 33.7 | 139.6 | 170.0 | 121.3 | 15.0 | 14.1 | 164.4 | 130.1 | 129.8 | 134.5 | 133.4 | 129.7 | 128.0 |
| 2a | 43.8 | 31.3 | 132.5 | 207.3 | 54.9 | 81.9 | 44.8 | 26.5 | 34.3 | 34.1 | 139.9 | 170.8 | 121.5 | 15.8 | 14.6 | 132.9 | 161.9 | 116.1 | 132.6 | 164.2 | 132.6 | 115.9 |
| 2b | 43.6 | 31.3 | 134.7 | 207.8 | 54.9 | 81.8 | 44.8 | 26.5 | 34.2 | 34.0 | 140.0 | 170.3 | 121.4 | 15.7 | 14.5 | 132.5 | 137.6 | 126.7 | 162.8 | 116.4 | 130.4 | 116.7 |
| 2c | 43.6 | 31.4 | 135.5 | 207.5 | 54.9 | 81.8 | 44.8 | 26.5 | 34.3 | 34.1 | 140.1 | 170.3 | 121.3 | 15.8 | 14.5 | 125.7 | 123.6 | 160.4 | 115.9 | 131.3 | 124.2 | 130.1 |
| 2d | 43.6 | 31.3 | 134.7 | 207.7 | 54.9 | 81.8 | 44.8 | 26.5 | 34.2 | 34.0 | 140.0 | 170.3 | 121.4 | 15.7 | 14.5 | 129.5 | 134.8 | 130.0 | 137.2 | 130.1 | 132.2 | 128.8 |
| 2e | 43.7 | 31.4 | 134.1 | 207.8 | 54.9 | 81.8 | 44.8 | 26.5 | 34.2 | 34.1 | 140.0 | 170.3 | 121.4 | 15.8 | 14.5 | 132.6 | 134.1 | 131.9 | 132.1 | 124.0 | 132.1 | 131.9 |
| 2f | 43.6 | 31.2 | 134.8 | 207.7 | 54.8 | 81.7 | 44.7 | 26.4 | 34.2 | 34.0 | 140.0 | 170.2 | 121.3 | 15.7 | 14.4 | 132.1 | 137.5 | 132.9 | 122.9 | 132.4 | 130.3 | 129.1 |
| 2g | 43.7 | 31.0 | 135.8 | 207.4 | 54.9 | 81.8 | 44.7 | 26.3 | 34.0 | 34.0 | 140.0 | 170.3 | 121.2 | 15.7 | 14.4 | 132.2 | 135.2 | 126.2 | 133.4 | 130.5 | 127.3 | 130.1 |
| 2h | 43.7 | 31.5 | 132.9 | 208.1 | 54.8 | 81.9 | 44.9 | 26.6 | 34.3 | 34.2 | 140.2 | 170.3 | 121.2 | 15.8 | 14.6 | 134.0 | 132.4 | 130.8 | 128.4 | 146.3 | 128.4 | 130.8 |
| 2i | 43.7 | 31.5 | 132.5 | 208.1 | 54.8 | 81.9 | 44.9 | 26.6 | 34.3 | 34.2 | 140.2 | 170.3 | 121.3 | 15.8 | 14.6 | 134.0 | 133.1 | 130.8 | 127.0 | 150.9 | 127.0 | 130.8 |
| 3a | 43.5 | 31.3 | 137.9 | 207.1 | 55.0 | 81.7 | 44.7 | 26.3 | 34.0 | 34.0 | 139.9 | 170.3 | 121.4 | 15.7 | 14.4 | 124.0 | 124.4 | 159.7 | 162.3 | 132.7 | 121.1 | 138.8 |
| 3b | 43.8 | 31.6 | 131.6 | 209.8 | 54.9 | 82.1 | 45.0 | 26.7 | 34.2 | 34.3 | 140.2 | 170.7 | 121.5 | 15.9 | 14.7 | 129.9 | 119.9 | 156.7 | 117.1 | 142.6 | 121.3 | 129.5 |
| 3c | 43.8 | 31.1 | 131.6 | 208.4 | 54.8 | 82.1 | 44.8 | 26.4 | 34.0 | 34.2 | 139.9 | 170.8 | 121.7 | 15.8 | 14.5 | 133.1 | 117.6 | 129.2 | 126.7 | 156.6 | 117.6 | 135.9 |
| 3d | 43.8 | 31.1 | 133.5 | 208.5 | 54.9 | 82.1 | 44.7 | 26.4 | 34.1 | 34.0 | 139.9 | 170.7 | 121.6 | 15.7 | 14.4 | 128.0 | 124.2 | 155.9 | 117.3 | 130.7 | 124.0 | 128.8 |
| 4a | 43.1 | 30.0 | 130.7 | 207.7 | 54.9 | 81.8 | 44.7 | 26.4 | 34.1 | 33.9 | 140.0 | 170.2 | 121.1 | 15.6 | 14.4 | 119.9 | 152.1 | - | 145.1 | 112.5 | 116.4 | - |
| 4b | 43.8 | 31.5 | 133.6 | 208.0 | 54.9 | 81.9 | 44.9 | 26.6 | 34.3 | 34.2 | 140.2 | 170.3 | 121.3 | 15.9 | 14.2 | 134.1 | - | - | - | - | - | - |
| 5a | 46.2 | 24.6 | 35.3 | 221.7 | 55.0 | 82.3 | 45.9 | 18.4 | 37.4 | 35.4 | 45.8 | 174.8 | 28.6 | 16.9 | 16.1 | - | - | - | - | - | - | - |
| 5b | 46.8 | 24.3 | 36.9 | 219.3 | 55.1 | 83.1 | 45.2 | 25.3 | 33.5 | 34.8 | 47.2 | 177.6 | 33.3 | 16.2 | 14.3 | - | - | - | - | - | - | - |
In 1d, the acetyl signals appear at 20.7 and 170.0 ppm.
In 4b, the naphthyl signals appear at 126.8, 127.0, 127.4, 127.8, 128.5, 128.7, 131.4, 133.0, 133.3 and 133.6 ppm.
In 5a, the cysteine signals appear at 38.3, 54.8, 174.2 and 52.5 ppm.
In 5b, the cysteine signals appear at 38.2, 54.8, 176.4 and 52.7 ppm.
Table 3: 13C NMR chemical shifts (in ppm) for the assayed compounds 1a-1f, 2a-2i, 3a-3d, 4a-4b and 5a-5b determined at 100 MHz in CDCl3.
In 4b, the naphthyl signals appear at 126.8, 127.0, 127.4, 127.8, 128.5, 128.7, 131.4, 133.0, 133.3 and 133.6 ppm.
In 5a, the cysteine signals appear at 38.3, 54.8, 174.2 and 52.5 ppm.
In 5b, the cysteine signals appear at 38.2, 54.8, 176.4 and 52.7 ppm.
Table 3: 13C NMR chemical shifts (in ppm) for the assayed compounds 1a-1f, 2a-2i, 3a-3d, 4a-4b and 5a-5b determined at 100 MHz in CDCl3.
The MCF-10A cell line used in this study is a non-tumorigenic breast epithelial cell line [23] while the JIMT-1 cells are breast cancer-derived tumorigenic. [24] Table 1 shows the experimental cytotoxicity IC50 values in mM obtained with the two cell lines, calculated from the dose response curves. In addition, the ratios between the IC50 MCF-10A and IC50 JIMT-1 were calculated, and are presented in Table 1 as a measure for the selectivity of each compound. The IC50 values show consistently that the JIMT-1 cells are more sensitive to the compounds assayed compared to the MCF-10A cells, which is consistent with our previous observation. [21] A cellular component that has been recognized to be able to neutralize SLs is glutathione. [25] Glutathione can react spontaneously with various electrophilic substrates, e.g. Michael acceptors, but it is also conjugated to xenobiotics in a reaction catalyzed by glutathione S-transferase. [26] It has been shown that the reaction of glutathione with SLs is reversible. [27] When measuring glutathione levels in MCF-10A and JIMT-1 cells, we found that the former have a 10 times higher concentration than the latter. In addition, the JIMT-1 cells have low expression of the glutathione S-transferase M1 gene. [24] Our notion that the difference in sensitivity between the normal-like MCF-10A and the JIMT-1 breast cancer cells is due to the lower glutathione concentration and low expression of the glutathione S-transferase gene in the latter needs further experimental clarification.
It is interesting that the oxidized derivatives (1b-1f) of 1a are less potent, especially the 3-acylated derivatives (1d-1f). Compared to the 3-bezylidenes, carbon 3 of 1c-1f is chiral and the substituent is directed in a certain direction. This may be disadvantageous for the ability of 1c-1f to react with certain critical nucleophilic sites in the target proteins. Besides this observation, it can be noted that the benzylidene derivatives 2a-2i and 3a-3d show the expected potency in the JIMT-1 cells, but do not show a promising selectivity. The derivatives 5a and 5b, which formally lack the α-methylene-β-lactone moiety, are still cytotoxic. Presumably this is due to the reversibility of the Michael reaction, which would cause the derivatives 5a and 5b to regenerate damsin (1a) during the experiment. It would consequently be of interest to measure the reversibility of this reaction during relevant conditions. Another biological explanation to the difference in toxicity between 5a and 5b could be that the toxicity of SLs is not only dependent on the presence of an a-methylene-g-lactone group but that there may be other toxic mechanisms independent of binding to p65/NF-kB [28,29] and this notion needs investigation as well. The stereoisomers 5a and 5b show slightly different toxicities (see Table 1) which may be related to different binding affinities to possible targets in the cell and/or differences in chemical stability.
In conclusion, with this paper and our previous paper [21], we demonstrate the synthesis of 43 unique damsin derivatives, many of which that show toxicity with a lower IC50 than that found for damsin itself. Most important is the increase in selective toxicity i.e. a lower IC50 in the JIMT-1 cancer cell line compared to the MCF-10A cell line. Further studies are needed to unravel the role of different chemical and biological entities that define toxicity with the goal to increase the selectivity towards cancer cells.
Supporting Information
Compound 1a (damsin)
1H NMR data (400 MHz, CDCl3): 6.13 (1H, s, H-13b), 5.46 (1H, s, H-13b), 4.44 (1H, d, J 6.9 Hz, H-6), 3.24 (1H, m, H-7), 2.38 (2H, m, H-3), 2,15 (1H, m, H-10), 2,01 (1H, m, H-1) 1,95 (1H, m, H-8b), 1,81 (2H, m, H-9a,b), 1,79 (1h, m, H-8a), 1,69 (2H, m, H-2), 1,01 (3H, d, J 7.5 Hz, H-14), 1,0 (3H, m, H-15). 13C NMR data (100 MHz, CDCl3): 218.9 (C-4), 170.1 (C-12), 139.5 (C-11), 120.6 (C-13), 81.6 (C-6), 54.7 (C-5), 45.8 (C-1), 44.2 (C-7), 36.0 (C-3), 34.1 (C-10), 33.2 (C-9), 25.5 (C-2), 23.8 (C-8), 15.7 (C-14), 13.6 (C-15).
Compound 1b (coronopilin)
1H NMR data (400 MHz, CDCl3): 6.26 (1H, s, H-13b), 5.58 (1H,s, H-13a), 4.92 (1H,d, J 8.2 Hz, H-6), 3.34 (1H, m, H-7), 2.65 (1H, dd, J 23.2, 10.5 Hz, H-3b), 2.47 (2H, m, H-2), 2,40 (1H, m, H-9b), 2,19 (1H, m, H-10), 2,08 (1H, m, H-8b), 1,71(1H, m, H-8a), 1,61 (1H, m, H-3a), 1,60 (1H, m, H-9a), 1.20 (3H, d, J 7.5 Hz, H-14), 1.15 (3H, s, H-15). 13C NMR data (100 MHz, CDCl3): 216.4 (C-4), 169.4 (C-12), 140.0 (C-11), 120.7 (C-13), 84.2 (C-1), 78.4 (C-6), 57.9 (C-5), 43.9 (C-7), 42.1 (C-10), 32.3 (C-2), 31.0 (C-3), 29.4 (C-9), 26.8 (C-8), 16.5 (C-14), 13.9 (C-15).
Compound 1c (3α-hydroxydamsin)
1H NMR data (400 MHz, CDCl3): 2.35 (1H, ddd, J 12.3; 7.6; 4.5 Hz, H-1), 1.81 (1H, m, H-2a), 2.48 (1H, ddd, J 14.4; 12.6; 9.7 Hz, H-2b), 5.19 (1H, dd, J 9.6; 2.4 Hz, H-3a), 4.70 (1H, d, J 8.9, H-6), 3.31 (1H, m, H-7), 1.86 (1H, m, H-8a), 1.99 (1H, m, H-8b), 1.70 (1H, m, H-9a), 1.87 (1H, m, H-9b), 2.14 (1H, m, H-10), 5.55 (1H, d, J 2.8Hz, H13a), 6.26 (1H, d, J 3.1Hz, H-13b), 1.07 (3H, d, J 7.5 Hz, CH3-14), 1.12 (3H, s, CH3-15), 2.08 (3H, s, CH3-17). 13C NMR data (100 MHz, CDCl3): 42.6 (C-1), 30.1 (C-2), 71.5 (C3), 211.8 (C-4), 55.2 (C-5), 81.3 (C-6), 44.0 (C-7), 25.9 (C-8), 34.0 (C-9), 33.7 (C-10), 139.7 (C-11), 170.0 (C-12), 121.3 (C-13), 15.0 (C-14), 14.1 (C15), 170.0 (C-16), 20.7 (C-17). [α]D20 -5 (c 1.00, CH2Cl2). IR spectrum (film, ν, cm-1 ): 2928, 2870, 1753, 1736, 1659, 1451, 1372, 1337, 1271, 1223, 1164, 1113, 1059, 1006, 977, 948, 882, 815, 733, 700, 627, 529, 460. HRMS-ESI (m/z, %) 265.1451 (100) [M+H]+ (calculated for C15H21O4 265.1440).
Compound 1d (3α-acetoxydamsin)
1H NMR data (400 MHz, CDCl3): 2.35 (1H, ddd, J 12.3; 7.6; 4.5 Hz, H-1), 1.81 (1H, m, H-2a), 2.48 (1H, ddd, 3 J 14.4; 12.6; 9.7 Hz, H-2b), 5.19 (1H, dd, J 9.6; 2.4 Hz, H-3a), 4.70 (1H, d, J 8.9 Hz, H-6), 3.31 (1H, m, H-7), 1.86 (1H, m, H-8a), 1.99 (1H, m, H-8b), 1.70 (1H, m, H-9a), 1.87 (1H, m, H-9b), 2.14 (1H, m, H-10), 5.55 (1H, d, J 2.8 Hz, H-13a), 6.26 (1H, d, J 3.1Hz, H-13b), 1.07 (3H, d, J 7.5, CH3-14), 1.12 (3H, s, CH3-15), 2.08 (3H, s, CH3-17). 13C NMR data (100 MHz, CDCl3): 42.6 (C-1), 30.1 (C-2), 71.5 (C3), 211.8 (C-4), 55.2 (C-5), 81.3 (C-6), 44.0 (C-7), 25.9 (C-8), 34.0 (C-9), 33.7 (C-10), 139.7 (C-11), 170.0 (C-12), 121.3 (C-13), 15.0 (C-14), 14.1 (C15), 170.0 (C-16), 20.7 (C-17). [α]D20 -20.5 (c 1.00, CH2Cl2). IR spectrum (film, ν, cm-1 ): 2928, 2870, 1753, 1736, 1659, 1451, 1372, 1337, 1271, 1223, 1164, 1113, 1059, 1006, 977, 948, 882, 815, 733, 700, 627, 529, 460. HRMS-ESI (m/z, %) 307.1549 (100) [M+H]+ (calculated for C17H23O5 (307.1546).
Compound 1e (3α-benzoyloxydamsin)
1H NMR data (400 MHz, CDCl3): 8.00 (2H, dd, J 8.4; 1.3 Hz, H-2´´/6´´), 7.56 (1H, m, H-4´), 7.42 (2H, m, 3´´/5´´), 6.28 (1H, d, J 3.2 Hz, H-13b), 5.57 (1H, d, J 2.8 Hz, H-13a), 5.39 (1H, dd, J 9.5, 2.4 Hz, H-3), 4.79 (1H, d, J 8.8 Hz, H-6), 3.34 (1H, m, H-7), 2.59 (1H, m, H-2b), 2.49 (1H, m, H-1), 2.16 (1H, m, H-10), 1.86 (2H, m, H-8a), 1.73 (1H, m, H-9a), 1.18 (3H, s, CH3-15), δ 1.10 (3H, d, J 7.5 Hz, CH3-14); 13C NMR data (100 MHz, CDCl3): 211.8 (C-4), 170.1 (C-12), 165.7 (C-1´), 133.5 (C-4´´), 129.9 (C-2´´/6´´), 129.3 (C-1´´), 128.5 (C-3´´/5´´), 121.4 (C-13), 81.5 (C-6), 72.2 (C-3), 55.4 (C-5), 44.1 (C-7), 42.8 (C-1), 34.0 (C-9), 33.8 (C-10), 30.3 (C-2), 26.0 (C-8), 15.2 (C-14), 14.3 (C-15); [α]D20 -29.5 (c 1.00, CH2Cl2); IR spectrum (film, γ, cm-1): 2963, 1755, 1719, 1601, 1473, 1450, 1389, 1343, 1314, 1262, 1231, 1196, 1175, 1152, 1111, 1068, 1021, 975, 935, 892, 801, 708, 687, 642, 584, 534; TOFMS: 369.1701 [M+H+] (calculated for C22H25O5 369.1702).
Compound 1f (3α-(m-chlorobenzoyloxy) damsin)
1H NMR data (400 MHz, CDCl3): 7.96 (1H, t, J 1.7; 1.7 Hz, H-2´´), 7.88 (1H, dt, J 7.8; 1.2; 1.2 Hz, H-6´´), 7.53 (1H, dq, J 7.9; 1.1;1.1; 1.1 Hz, H-4´´), 7.36 (1H, t, J 7.9; 7.9 Hz, H-5´´), 6.27 (1H, d, J 3.1 Hz, H-13b), 5.56 (1H, d, J 2.8 Hz, H-13a), 5.42 (1H, dd, J 9.8; 2.4 Hz, H-3a), 4.78 (1H, d, J 8.9 Hz, H-6), 3.35 (1H, m, H-7), 2.59 (1H, ddd, J 14.5; 12.6; 9.9 Hz, H-2b), 2.46 (1H, ddd, J 12.3; 7.7; 4.4 Hz, H-1), 2.17 (1H, m, H-10), 1.99 (1H, m, H-8b), 1.96 (1H, m, H-2a), 1.88 (1H, m, H-8a), 1.87 (1H, m, H-9b), 1.72 (1H, m, H-9a), 1.17 (3H, s, CH3-15), 1.10 (3H, d, J 7.5 Hz, CH3-14); 13C NMR data (100 MHz, CDCl3): 211.3 (C-4), 170.0 (C-12), 164.4 (C-1´), 139.6 (C-11), 134.5 (C-3´´), 133.4 (C-4´´), 130.1 (C-1´´), 129.8 (C-2´´), 129.7 (C-5´´), 128.0 (C-6´´), 121.3 (C-13), 81.3 (C-6), 72.3 (C-3), 55.3 (C-5), 44.0 (C-7), 42.7 (C-1), 33.9 (C-9), 33.7 (C-10), 30.0 (C-2), 25.9 (C-8), 15.0 (C-14), 14.1 (C-15). [α]D20 -40.0 (c 1.00, CH2Cl2). IR spectrum (film, γ, cm-1): 3071, 2960, 2936, 2873, 1756, 1722, 1661, 1574, 1473, 1451, 1426, 1388, 1364, 1336, 1277, 1254, 1164, 1126, 1087, 1073, 1009, 978, 903, 812, 747, 736, 700, 674, 632; TOFMS: 403.1325 [M+H+] (calculated for C22H24ClO5 403.1312).
Compound 2a [(E)-3-(p-(fluorophenyl) methylene) damsin]
1H NMR data (400 MHz, CDCl3): 7.54 (2H, m, H-5´´/3´´ ), 7.40 (1H, m, H-1´), 7.11 (2H, m, H-2´´/6´´), 6.29 (1H, d, J 3.0 Hz, H-13b), 5.58 (1H, d, J 2.7 Hz, H-13a), 4.65 (1 H, d, J 8.5 Hz, H-6), 3.29 (1H, m, H-7), 2.94 (1H, ddd, J 15.7; 12,3; 3.1 Hz, H-2b), 2.83 (1H, ddd, J 16.8; 7.3; 1.9 Hz, H-2a), 2.28 (1H, m, H-10), 2.08 (1H, m, H-8b), 2.07 (1H, m, H-1), 1.87 (1H, m, H-9b), 1.78 (1H, m, H-8a), 1.74 (1H, m, H-9a), 1.17 (3H, d, J 7.5 Hz, CH3-14), 1.16 (3H, s, CH3-15); 13C NMR data (100 MHz, CDCl3): 207.3 (C-4), 170.8 (C-12), 164.2 (C-4´´), 161.9 (C-1´´), 139.9 (C-11), 132.9 (C-1´), 132.6 (C-3´´/5´´), 132.5 (C-3), 121.5 (C-13), 116.1 (C-2´´), 115.9 (C-6´´), 81.9 (C-6), 54.9 (C-5), 44.8 (C-7), 43.8 (C-1), 34.3 (C-9), 34.1 (C-10), 31.3 (C-2), 26.5 (C-8), 15.8 (C-14), 14.6 ( C-15). [α]D20 -9.5 (c 1.00, CH2Cl2); IR spectrum (film, γ, cm-1). HRMS-ESI (m/z) 355.1702 [M+H]+ (calculated for C22H24FO3 355.1710).
Compound 2b [(E)-3-(m-(fluorophenyl) methylene) damsin]
1H NMR data (400 MHz, CDCl3): 7.39 (1H, m, H-5´´), 7.37 (1H, m, H-1´), 7.30 (1 H, d, J 7.8 Hz, H-2´´), 7.23 (1H, dd, J 9.9; 2.0 Hz, H-6´´), 7.06 (1H, m, H-4´´), 6.28 (1H, d, J 3.0 Hz, H-13b), 5.57 (1H, d, J 2.7 Hz, H-13b), 4.64 (1H, d, J 8.5 Hz, H-6), 3.30 (1H, tdd, J 11.3; 5.4; 2.9 Hz, H-7), 2.95 (1H, ddd, J 16.1; 12.6; 3.2 Hz, H-2b), 2.85 (1H, ddd, J 17.0; 7.4; 2.0 Hz, H-2a), 2.28 (1H, m, H-10), 2.10 (1H, m, H-1), 2.07 (1H, m, H-8b), 1.90 (1H, m, H-9b), 1.81 (1H, m, H-8a) 1.72 (1H, m, H-9a), 1.18 (3H, d, J 7.5 Hz, CH3-14), 1.15 (3H, s, CH3-15); 13C NMR data (100 MHz, CDCl3): 207.8 (C-4), 170.3 (C-12), 162.8 (C-3´´), 140.0 (C-11), 137.6 (C-1´´), 134.7 (C-3), 132.5 (C-1´), 130.4 (C-5´´) 126.7 ( C-2´´), 121.4 (C-13), 116.7 (C-6´´), 116.4 (C-4´´), 81.8 (C-6), 54.9 (C-5), 44.8 (C-7), 43.6 (C-1), 34.2 (C-9), 34.0 (C-10), 31.3 (C-2), 26.5 (C-8), 15.7 (C-14), 14.5 (C-15); [α]D20 +4.5 (c 1.00, CH2Cl2); IR spectrum (film, γ, cm-1): 2926, 2869, 1757, 1716, 1629, 1580, 1485, 1444, 1385, 1336, 1272, 1230, 1210, 1189, 1160, 1118, 1079, 1058, 1003, 990, 952, 884, 865, 814, 788, 735, 682. HRMS-ESI (m/z) 355.1723 [M+H]+ (calculated for C22H24FO3 355.1710).
Compound 2c [(E)-3-(o-(fluorophenyl) methylene) damsin]
1H NMR data (400 MHz, CDCl3): 7.62 (1 H, s, H-1´), 7.53 (1H, td, J 7.7; 1.6 Hz, H-6´´), 7.35 (1H, m, H-4´´), 7.18 ( 1H, td, J 7.6; 0.8 Hz, H-5´´), 7.09 (1H, m, H-3´´), 6.27 (1H, d, J 3.0 Hz, H-13b), 5.57 (1H, d, J 2.7 Hz, H-13a), 4.64 (1H, d, J 8.5 Hz, H-6), 3.29 (1H, m, H-7), 2.94 (1H, ddd, J 16.2; 12.6; 3.3 Hz, H-2b), 2.76 (1H, ddd, J 16.9; 7.1; 1.8 Hz, H-2a), 2.26 (1H, m, H-10), 2.07 (1H, m, H-8b), 2.06 (1H, m, H-1), 1.86 (1H, m, H-9b), 1.82 (1H, m, H-8a), 1.73 (1H, m, H-9a), 1.17 (3H, d, J 7.5 Hz, CH3-14), 1.16 (3H, s, CH3-15); 13C NMR data (100 MHz, CDCl3): 207.6 (C-4), 170.3 (C-12), 162.9-160.4 (C-2´´), 140.1 (C-11), 135.5 (C-3), 131.3 (C-4´´), 130.1 (C-6´´), 125.7 (C-1´), 124.2 (C-5´´), 123.6 (C-1´´), 121.3 (C-13), 116.17 (s), 115.9 (C-3´´), 81.8 ( C-6), 54.9 (C-5), 44.8 (C-7), 43.6 (C-1), 34.1 (C-10), 31.4 (C-2), 26.5 (C-8), 15.8 (C-14), 14.5 (C-15) ; [α]D20 +9.8 (c 1.00, CH2Cl2); IR spectrum (film, γ, cm-1): 2925, 2857, 1757, 1716, 1627, 1610, 1484, 1453, 1385, 1336, 1271, 1254, 1229, 1211, 1194, 1162, 1118, 1058, 985, 951, 913, 884, 838, 814, 799, 760. HRMS-ESI (m/z) 355.1716 [M+H]+ (calculated for C22H24FO3 355.1710).
Compound 2d [(E)-3-(m-(chlorophenyl) methylene) damsin]
1H NMR data (400 MHz, CDCl3): 7.50 (1 H, s, H-2´´), 7.40 (1H, m, H-6´´), 7.35 (1H, m, H-4´´), 7.34 (1H, m, H-5´´), 7.33 (1H, m, H-1´), 6.28 (1H, d, J 3.0 Hz, H-13b) 5.57 (1H, d, J 2.7 Hz, H-13a), 4.64 (1H, d, J 8.5 Hz, H-6), 3.30 (1H, m, H-7), 2.95 (1H, ddd, J 16.6; 12.3; 3.2 Hz, H-2b), 2.84 (1H, ddd, J 17.0; 7.3; 2.0 Hz, H-2a), 2.29 (1H, m, H-10), 2.08 (1H, m, H-1), 2.06 (1H, m, H-8b), 1.91 (1H, m, H-9b), 1.82 (1H, m, H-8a), 1.73 (1H, m, H-9a), 1.18 (3H, d, J 7.5 Hz, CH3-14), 1.15 (3H, s, CH3-15); 13C NMR data (100 MHz, CDCl3): 207.7 (C-4), 170.3 (C-12), 140.0 (C-11), 137.2 (C-3´´), 134.8 (C-1´´) 134.7 (C-3), 132.2 (C-5´´), 130.1 (C-4´´), 130.0 (C-2´´), 129.5 (C-1´), 128.8 (C-6´´), 121.4 (C-13), 81.8 (C-6) 54.9 (C-5), 44.8 (C-7), 43.6 (C-1), 34.2 (C-9), 34.0 (C-10), 31.3 (C-2), 26.5 (C-8), 15.7 (C-14), 14.5 (C-15); [α]D20 -8.2 (c 1.00, CH2Cl2); IR spectrum (film, γ, cm-1): 2925, 2866, 1758, 1716, 1628, 1562, 1473, 1449, 1420, 1385, 1337, 1271, 1250, 1206, 1186, 1160, 1118, 1058, 987, 952, 886, 815, 787, 734, 683. HRMS-ESI (m/z) 371.1425 [M+H]+ (calculated for C22H24ClO3 371.1414).
Compound 2e [(E)-3-(p-bromophenylmethylene) damsin]
1H NMR data (400 MHz, CDCl3): 7.55 (2H, m, H-3´´/5´´), 7.4 (2H, d, J 6.8 Hz, H-2´´/6´´), 7.36 (1H, m, H-1´), 6.28 (1H, dd, J 6.7, 3.1 Hz, H-13b), 5.56 (1H, dd, J 8.5; 2.7 Hz, H-13a), 4.65 (1H, m, H-6), 3.29 (1H, m, H-7), 2.93 (1H, ddd, J 16.8, 12.3, 3.2 Hz, H-2b), 2.81 (1H, ddd, J 16.9; 7.3; 2.0 Hz, H-2a), 2.28 (1H, m, H-10), 2.09 (1H, m, H-8b), 2.08 (1H, m, H-1), 1.90 (1H, m, H-9b), 1.83 (1H, m, H-8a), 1.75 (1H, m, H-9a), 1.18 (3H, d, J 7.5 Hz, CH3-14), 1.16 (3H, s, CH3-15); 13C NMR data (100 MHz, CDCl3): 207.8 (C-4), 170.3 (C-12), 140.0 (C-11), 134.4 (C-3), 134.1 (C-1´´), 132.6 (C-1´), 132.1 (C-3´´/5´´), 131.9 (C-2´´/6´´), 124.0 (C-4´´), 121.4 (C-13), 81.8 (C-6), 54.9 (C-5), 44.8 (C-7), 43.7 (C-1), 34.2 (C-9), 34.1 (C-10), 31.4 (C-2), 26.5 (C-8), 15.8 (C-14), 14.5 (C-15) ; [α]D20 +24.5 (c 1.00, CH2Cl2); IR spectrum (film, γ, cm-1): 2925, 2861, 1757, 1714, 1624, 1583, 1487, 1448, 1403, 1337, 1306, 1271, 1252, 1238, 1161, 1119, 1073, 1006, 984, 951, 815, 733. HRMS-ESI (m/z) 415.0917 [M+H]+ (calculated for C22H24BrO3 415.0909).
Compound 2f [(E)-3-(m-bromophenylmethylene) damsin]
1H NMR data (400 MHz, CDCl3): 7.64 (1H, s, H-2´´), 7.48 (1H, d, J 8.0 Hz, H-4´´), 7.44 (1H, d, J 7.9 Hz, H-6´´), 7.31 (1H, m, H-1´), 7.28 (1H, t, J 7.9 Hz, H-5´´), 6.27 (1H, d, J 3.0 Hz, H-13b), 5.57 (1H, d, J 2.6 Hz, H-13a), 4.63 (1H, d, J 8.6 Hz, H-6), 3.29 (1H, m, H-7), 2.94 (1H, ddd, J 16.6; 12.3; 3.1 Hz, H-2b), 2.83 (1H, ddd, J 17.0; 7.3; 1.9 Hz, H-2a), 2.29 (1H, m, H-10), 2.08 (1 H, m, H-1), 2.07 (1H, m, H-8b), 1.89 (1H, m, H-9b), 1.83 (1H, m, H-8a), 1.74 (1H, m, H-9a), 1.17 (3H, d, J 7.5 Hz, CH3-14), 1.14 (3H, s, CH3-15); 13C NMR data (100 MHz, CDCl3): 207.7 (C-4), 170.2 (C-12), 140.0 (C-11), 137.5 (C1´´), 134.8 (C-3), 132.9 (C-2´´), 132.4 (C-4´´), 132.1 (C-1´), 130.3 (C-5´´), 129.1 (C-6´´), 122.9 (C-3´), 121.3 (C-13), 81.7 (C-6), 54.8 (C-5), 44.7 (C-7), 43.6 (C-1), 34.2 (C-9), 34.0 (C-10), 31.2 (C-2), 26.4 (C-8), 15.7 (C-14), 14.4 (C-15) ; [α]D20 -2.6 (c 1.00, CH2Cl2); IR spectrum (film, γ, cm-1): 2923, 2860, 1757, 1716, 1627, 1557, 1473, 1449, 1415, 1337, 1270, 1160, 1118, 986, 785, 682. . HRMS-ESI (m/z) 415.0919 [M+H]+ (calculated for C22H24BrO3 415.0909).
Compound 2g [(E)-3-(o-bromophenylmethylene) damsin]
1H NMR data (400 MHz, CDCl3): 7.67 (1H, dd, J 2.9; 1.9 Hz, H-1), 7.61 (1H, dd, J 8.0, 1.1 Hz, H-3´´), 7.50 (1H, dd, J 7.8; 1.4 Hz, H-6´´), 7.34 (1H, m, H-5´´), 7.19 (1H, td, J 7.8, 1.6 Hz, H-4´´), 6.26 (1H, d, J 3.0 Hz, H-13b), 5.56 (1H, d, J 2.7 Hz, H-13a), 4.61 (1H, d, J 8.5 Hz, H-6), 3.29 (1H, ddd, J 10.4; 7.6; 3.2 Hz, H-7), 2.90 (1H, ddd, J 16.3; 12.7; 3.4 Hz, H-2b), 2.69 (1H, ddd, J 16.7; 6.9; 1.8 Hz, H-2a), 2.21 (1H, ddd, J 11.7; 7.7; 4.3 Hz, H-10), 2.06 (1H, m, H-8b), δ 2.04 (1H, m, H-1), 1.85 (1H, m, H-9b), 1.81 (1H, m, H-8a), 1.72 (1H, m, H-9a), 1.16 (3H, s, CH3-15), 1.14 (3H, d, 7.5 Hz, CH3-14); 13C NMR data (100 MHz, CDCl3): 207.4 (C-4), 170.3 (C-12), 140.0 (C-11), 135.8 (C-3), 135.2 (C-1´´), 133.4 (C-3´´), 132.2 (C-1´), 130.5 (C-4´´), 130.1 (C-6´´), 127.3 (C-5´´), 126.2 (C-2´´), 121.2 (C-13), 81.8 (C-6), 54.9 (C-5), 44.7 (C-7), 43.7 (C-1), 34.0 (C-9), 34.0 (C-10), 31.0 (C-2), 26.3 (C-8), 15.7 (C-14), 14.4 (C-15); [α]D20 +9.1 (c 1.00, CH2Cl2); IR spectrum (film, γ, cm-1): 2924, 2865, 1758, 1717, 1625, 1465, 1432, 1385, 1336, 1271, 1251, 1227, 1188, 1161, 1113, 1046, 1023, 985, 950, 815, 761, 740, 664. HRMS-ESI (m/z) 415.0912 [M+H]+ (calculated for C22H24BrO3 415.0909).
Compound 2h [(E)-3-(p-ethylphenylmethylene) damsin]
1H NMR data (400 MHz, CDCl3): 7.47 (2H, d, J 8.2 Hz, H-2´´/6´´), 7.42 (1H, m, H-1´), 7.25 (2H, d, J 7.9 Hz, H-3´´/5´´), 6.28 (1H, d, J 2.9 Hz, H-13b), 5.57 (1H, d, J 2.6 Hz, H-13a), 4.65 (1H, d, J 8.5 Hz, H-6), 3.28 (1H, m, H-7), 2.96 (1H, ddd, J 16.8; 7.4, 1.9 Hz, H-2b), 2.86 (1H, ddd, J 16.8; 7.4; 1.9 Hz, H-2a), 2.68 (2H, q, J 7.6 Hz, H-1´´´), 2.28 (1H, m, H-10), 2.08 (1H, m, H-8b), 2.07 (1H, m, H-1), 1.88 (1H, m, H-9b), 1.81 (1H, m, H-8a), 1.74 (1H, m, H-9a), 1.25 (3H, t, J 7.6 Hz, H-2´´´), 1.18 (3H, d, J 7.5 Hz, CH3-14), 1.16 (3H, s, CH3-15); 13C NMR data (100 MHz, CDCl3): 208.1 (C-4), 170.3 (C-12), 146.3 (C-4´´), 140.2 (C-11), 134.0 (C-1´), 132.9 (C-3), 132.4 (C-1´´), 130.8 (C-2´´/6´´), 128.4 (C-5´´/3´´), 121.2 (C-13), 81.9 (C-6), 54.8 (C-5), 44.9 (C-7), 43.7 (C-1), 34.3 (C-9), 34.2 (C-10), 31.5 (C-2), 28.9 (C-1´´´), 26.6 (C-8), 15.8 (C-14), 15.4 (C-2´´´), 14.6 (C-15); [α]D20 +38.3 (c 1.00, CH2Cl2); IR spectrum (film, γ, cm-1): 2963, 2928, 2869, 1757, 1713, 1624, 1605, 1510, 1451, 1385, 1335, 1271, 1254, 1159, 1117, 1058, 985, 915, 826. HRMS-ESI (m/z) 365.2109 [M+H]+ (calculated for C24H29O3 365.2117).
Compound 2i [(E)-3-(p-isopropylphenylmethylene) damsin]
1H NMR data (400 MHz, CDCl3): 7.49 (2H, d, J 8.3 Hz, H-2´´/6´´), δ 7.43 (1H, m, H-1´), δ 7.28 (2H, d, J 8.2 Hz, H-3´´/5´´), δ 6.28 (1H, d, J 3.0 Hz, H-13b), δ 5.57 (1H, d, J 2.6 Hz, H-13a), δ 4.65 (1H, d, J 8.5 Hz, H-6), δ 3.28 (1H, m, H-7), δ 2.96 (1H, ddd, J 16.3; 9.5; 3.6 Hz, H-2b), δ 2.93 (1H, m, (H3C)2HC-Ar), δ 2.87 (1H, ddd, J 16.8; 7.5; 2.0 Hz, H-2a), δ 2.28 (1H, m, H-10), δ 2.09 (1H, m, H-8b), δ 2.07 (1H, m, H-1), δ 1.88 (1 H, m, H-9b), δ 1.81 (1H, m, H-8a), δ 1.73 (1H, m, H-9a), δ 1.26 (6H, d, J 6.9 Hz, (H3C)2HC-Ar), δ 1.18 (3H, d, J 7.5 Hz, CH3-14), δ 1.16 (3H, s, CH3-15); 13C NMR data (100 MHz, CDCl3): 208.1 (C-4), 170.3 (C-12), 150.9 (C-4´´), 140.2 (C-11), 134.0 (C-1´), 133.1 (C-1´´), 132.5 (C-3), 130.8 (C-2´´/6´´), 127.0 (C-3´´/5´´), 121.3 (C-13), 81.9 (C-6), 54.8 (C-5), 44.9 (C-7), 43.7 (C-1), 34.3 (C-9), 34.2 (C-10), 34.1 ((H3C)2HC-Ar), 31.5 (C-2), 26.6 (C-8), 23.88 ((H3C)2HC-Ar), 15.8 (C-14), 14.6 (C-15); [α]D20 +40.2 (c 1.00, CH2Cl2); IR spectrum (film, γ, cm-1): 2959, 2926, 2868, 1759, 1714, 1624, 1605, 1458, 1417, 1384, 1362, 1336, 1309, 1271, 1254, 1160, 1118, 1055, 985, 825. HRMS-ESI (m/z) 379.2285 [M+H]+ (calculated for C25H31O3 379.2273).
Compound 3a [(E)-3-(o-(p-trifluoromethylfluorophenyl) methylene) damsin]
1H NMR data (400 MHz, CDCl3): 7.65 (1H, t, J 7.6 Hz, H-6´´), 7.55 (1H, s, H-1´), 7.45 (1H, d, J 8.2 Hz, H-5´´), 7.36 (1H, dd, J 9.4; 1.15 Hz, H-3´´),6.28 (1H, d, J 3.0 Hz, H-13b), 5.57 (1H, d, J 2.7 Hz, H-13a), 4.64 (1H, d, J 8.6 Hz, H-6), 3.31 (1H, m, H-7), 2.95 (1H, ddd, J 16.4; 12.7; 3.4 Hz, H-2b), 2.75 (1H, ddd, J 17.1; 7.0; 1.7 Hz, H-2a), 2.26 (1H, m, H-10), δ 2.09 (1H, m, H-1), 2.07 (1H, m, H-8b), 1.89 (1H, m, H-9b), 1.85 (1H, m, H-8a), 1.75 (1H, m, H-9a), 1.16 (3H, d, J 7.5 Hz, CH3-14), 1.16 (3H, s, CH3-15); 13C NMR data (100 MHz, CDCl3): 207.1 (C-4), 170.3 (C-12), 162.3 159.7 (C-2´´), 139.9 (C-11), 137.9 (C-3), 132.7 (C-4´´), 130.8 (C-6´´), 127.3 (C-1´´´), 124.4 (C-1´´), 124.0 (C-1´), 121.4 (C-13), 121.1 (C-5´´), 113.6 (C-3´´), 81.7 (C-6), 55.0 (C-5), 44.7 (C-7), 43.5 (C-1), 34.0 (C-9, 10), 31.3 (C-2), 26.3 (C-8), 15.7 (C-14), 14.4 (C-15); [α]D20 -18.6 (c 1.00, CH2Cl2); IR spectrum (film, γ, cm-1): 2928, 2868, 1758, 1720, 1636, 1574, 1507, 1426, 1329, 1272, 1210, 1195, 1167, 1122, 1066, 986, 949, 909, 880, 832, 815, 743, 659. HRMS-ESI (m/z) 423.1594 [M+H]+ (calculated for C23H23F4O3 423.1583).
Compound 3b [(E)-3-(o-(p-methylhydroxyphenyl) methylene) damsin]
1H NMR data (400 MHz, CDCl3): 8.0 (1H, s, H-1´), 7.36 (1 H, d, H-6´´), 6.73 (1H, d, H-5´´), 6.8 (1H, s, H-3´´), 6.3 (1H, d, J 3.0 Hz, H-13b), 5.58 (1H, d, J 2.7 Hz, H-13a), 4.65 (1H, d, J 8.4 Hz, H-6), 3.28 (1H, m, H-7), 2.93 (1H, ddd, J 16.7; 12.3; 3.1 Hz, H-2b), 2.8 (1H, ddd, J 16.6; 7.2; 1.7 Hz, H-2a), 2.32 (3H, s, H3C-Ar), 2.26 (1H, m, H-10), 2.07 (1H, m, H-8b), 2.06 (1H, m, H-1), 1.87 (1H, m, H-9b), 1.78 (1H, m, H-8a), 1.74 (1H, m, H-9a), 1.18 (3H, d, J 7.5 Hz, CH3-14), 1.17 (3H, s, CH3-14); 13C NMR data (100 MHz, CDCl3): 209.6 (C-4), 170.7 (C-12), 156.7 (C-2´´), 142.6 (C-4´´), 140.2 (C-11), 131.6 (C-3), 129.9 (C-1´), 129.5 (C-6´´), 121.5 (C-13), 121.3 (C-5´´), 119.9 (C-1´´), 117.1 (C-3´´), 82.1 (C-6), 54.9 (C-5), 45.0(C-7), 43.8 (C-1), 34.3 (C-10), 34.2 (C-9), 31.6 (C-2), 26.7 (C-8), 21.7(H3C-Ar), 15.9 (C-14), 14.7 (C-15); [α]D20 +45.6 (c 1.00, CH2Cl2); IR spectrum (film, γ, cm-1): 3349, 2924, 2858, 1742, 1710, 1658, 1605, 1450, 1419, 1384, 1342, 1305, 1261, 1187, 1162, 1107, 1058, 985, 884, 866, 814, 733, 643. HRMS-ESI (m/z) 367.1900 [M+H]+ (calculated for C23H27O4 367.1909).
Compound 3c [(E)-3-(p-(m-trifluoromethylhydroxyphenyl) methylene) damsin]
1H NMR data (400 MHz, CDCl3): 7.7 (1H, s, H-2´´), 7.52 (1H, d, H-6´´), 7.34 (1H, s, H-1´), 6.96 (1H, d, H-5´´), 6.27 (1H, d, J 3.0 Hz, H-13b), 5.57 (1H, d, J 2.7 Hz, H-13a), 4.63 (1H, d, J 8.5 Hz, H-6), 3.28 ( 1H, m, H-7), 2.91 (1H, ddd, J 15.5; 12.2; 3.1Hz, H-2b), 2.8 (1H, ddd, J 16.7; 7.4; 1.9 Hz, H-2a), 2.27 (1H, m, H-10), 2.07(1H, m, H-8b), 2.06 (1H, m, H-1), 1.89 (1H, m, H-9b), 1.84 (1H, m, H-8a), 1.73 (1H, m, H-9a), 1.16 (3H, d, J 7.5 Hz, CH3-14), 1.13 (3H, s, CH3-15); 13C NMR data (100 MHz, CDCl3): 208.4 (C-4), 170.8 (C-12), 156.6 (C-4´´), 139.9 (C-11), 135.9 (C-6´´), 133.1 (C-1´), 131.6 (C-3), 129.2 (C-2´´), 126.7 (C-3´´), 121.7 (C-13/ F3C-Ar), 117.6 (C-1´´/C-5´´), 82.1 (C-6), 54.8 (C-5), 44.8 (C-7), 43.8 (C-1), 34.2 (C-10), 34.0 (C-9), 31.1 (C-2), 26.4 (C-8), 15.8 (C-14), 14.5 (C-15); [α]D20 +16.4 (c 1.00, CH2Cl2); IR spectrum (film, γ, cm-1): 3273, 2925, 2856, 1737, 1713, 1606, 1509, 1435, 1365, 1327, 1297, 1274, 1252, 1207, 1185, 1159, 1123, 1052, 989, 952, 883, 832, 816, 756, 664. HRMS-ESI (m/z) 421.1632 [M+H]+ (calculated for C23H24F3O4 421.1627).
Compound 3d [(E)-3-(o-(m-chlorohydroxyphenyl) methylene) damsin]
1H NMR data (400 MHz, CDCl3): 7.71 (1H, m, H-1´), 7.34 (1H, d, J 2.5 Hz, H-2´´), 7.12 (1H, dd, J 8.7; 2.5 Hz, H-4´´), 6.77 (1H, d, J 8.7 Hz, H-5´´), 6.25 (1H, d, J 3.0 Hz, H-13b), 5.56 (1H, d, J 2.6 Hz, H-13a), 4.60 (1H, d, J 8.6 Hz, H-6), 3.26 (1H, m, H-7), 2.88 (1H, m, H-2b), 2.88 (1H, ddd, J 18.8; 7.1; 1.7 Hz, H-2a), 2.25 (1H, m, H-10), 2.04 (1H, m, H-8b), 2.03 (1H, m, H-1), 1.84 (1H, m, H-9b), 1.79 (1H, m, H-8a), 1.69 (1H, m, H-9a), 1.14 (3H, d, J 7.5 Hz, CH3-14), 1.12 (3H, s, CH3-15); 13C NMR data (100 MHz, CDCl3): 208.5 (C-4), 170.7 (C-12), 155.9 (C-6´´), 139.9 (C-11), 133.5 (C-3), 130.7 (C-4´´), 128.8 (C-2´´), 128.0 (C-1´), 124.2 (C-1´´), 124.0 (C-3´´), 121.6 (C-13), 117.3 (C-5´´), 82.1 (C-6), 54.9 (C-5) 44.7 (C-7) 43.8 (C-1), 34.1 (C-9), 34.0 (C-10), 31.1 (C-2), 26.4 (C-8), 15.7 (C-14), 14.4 (C-15); [α]D20 -7.0 (c 1.00, CH2Cl2); IR spectrum (film, γ, cm-1): 3343, 2924, 2856, 1738, 1713, 1656, 1615, 1490, 1474, 1447, 1417, 1385, 1342, 1274, 1255, 1192, 1159, 1115, 1058, 988, 951, 915, 896, 881, 816, 753, 665, 644. HRMS-ESI (m/z) 387.1370 [M+H]+ (calculated for C22H24ClO4 387.1363).
Compound 4a [(E)-3-(furanylmethylene) damsin]
1H NMR data (400 MHz, CDCl3): 7.53 (1H, d, J 1.4 Hz, H-3´´), 7.12 (1H, m, H-1´´), 6.63 (1H, d, J 3.5 Hz, .H-5´´), 6.47 (1 H, dd, J 3.4; 1.8 Hz, H-4´´) 6.20 (1H, d, J 3.0 Hz, H-13b), 5.51 (1H, d, J 2.7 Hz, H-13a), 4.59 (1H, d, J 8.5 Hz, H-6), 3.26 (1H, m, H-7), 2.94 (1H, ddd, J 17.8; 7.5; 1.8 Hz, H-2b), 2.82 (1H, m, H-2a), 2.23 (1H, m, H-10), 2.05 (1H, m, H-1), 2.01 (1H, m, H-8b), 1.82 (1H, m, H-9b), 1.78 (1H, m, H-8a), 1.68 (1H, m, H-9a), 1.10 (3H, d, J 7.5 Hz, CH3-14), 1.06 (3H, s, CH3-15); 13C NMR data (100 MHz, CDCl3): 207.7 (C-4), 170.2 (C-12), 152.1 (C-1´´), 145.1 (C-3´´), 140.0 (C-11), 130.7 (C-3), 121.1 (C-13), 119.9 (C-1´), 116.4 (C-5´´), 112.5 (C-4´´), 81.8 (C-6), 54.9 (C-5), 44.7 (C-7), 43.1 (C-1), 34.1 (C-9), 33.9 (C-10), 30.8 (C-2), 26.4 (C-8), 15.6 (C-14), 14.4 (C-15) ; [α]D20 -71.9 (c 1.00, CH2Cl2); IR spectrum (film, γ, cm-1): 2923, 2863, 1757, 1709, 1622, 1474, 1389, 1338, 1271, 1253, 1232, 1161, 1117, 1069, 1021, 982, 950, 882, 815, 752, 633. HRMS-ESI (m/z) 327.1584 [M+H]+ (calculated for C20H23O4 327.1596).
Compound 4b [(E)-3-(naphtylmethylene) damsin]
1H NMR data (400 MHz, CDCl3): 8.01 (1H, s, H-2´´), 7.89 (1H, m, H-4´´), 7.86 (1H, m, H-9´´), 7.83 (1H, m, H-7´´), 7.66 (1H, m, H-10´´), 7.60 (1H, m, H-1´), 7.53 (1H, m, H-6´´), 7.51 (1H, m, H-5´´), 6.30 (1H, J 3.0 Hz, H-13b), 5.58 (1H, J 2.6 Hz, H-13a). 4.67 (1H, J 8.5 Hz, H-6), 3.29 (1H, m, H-7), 3.08 (1H, ddd, J 16.2; 12.1; 3.2 Hz, H-2b), 2.97 (1H, ddd, J 16.8; 7.4; 1.9 Hz, H-2a), 2.32 (1H, m, H-10), 2.11 (1H, m, H-8b), 2.10 (1H, m, H-1), 1.91 (1H, m, H-9b), 1.82 ( 1H, m, H-8a), 1.76 (1H, m, H-9a), 1.22 (3H, d, J 7.5 Hz, CH3-14), 1.20 (3H, m, CH3-15); 13C NMR data (100 MHz, CDCl3): 208.0 (C-4), 170.3 (C-12), 140.2 (C-11), 134.1 (C-1´) 133.6 (C-3´´), 133.6 (C-3), 133.3 (C-8´´), 133.0 (C-1´´), 131.4 (C-2´´), 128.7 (C-9´´), 128.5 (C-4´´), 127.8 (C-7´´), 127.4 (C-6´´), 127.0 (C-10´´), 126.8 (C-5´´), 121.3 (C-13), 81.9 (C-6) 54.9 (C-5), 44.9 (C-7), 43.8 (C-1), 34.3 (C-9), 34.2 (C-10) 31.5 (C-2), 26.6 (C-8), 15.9 (C-14) 14.2 (C-15) ; [α]D20 +23.8 (c 1.00, CH2Cl2); IR spectrum (film, γ, cm-1): 2924, 2857, 1756, 1713, 1617, 1472, 1447, 1385, 1335, 1271, 1231, 1191, 1161, 1117, 1057, 988, 955, 882, 859, 816, 735, 640. HRMS-ESI (m/z) 387.1955 [M+H]+ (calculated for C26H27O3 387.1960).
Compound 5a
1H NMR data (400 MHz, CDCl3): 4.41 (1H, dd, J, 8.8; 2.8 Hz, H-6), 3.74 (1H, m, H-2´), 3.73 (3H, s, H-4´), 2.98 (1H, td, J 13.3; 4.8 Hz, H-1´b), 2.96 (1H, m, H-13b), 2.87 (1H, ddd, J 13.6; 6.6; 3.8 Hz, H-13a), 2.86 (1H, m, H-1b), 2.79 (1H, m, H-7), 2.78 (1H, ddd, J 18.7; 9.3; 4.5 Hz, H-7), 2.64 (1H, ddd, J 9.3; 6.3; 4.6 Hz, H-11), 2.40 (1H, m, H-3b), 2.22 (1H, m, H-3a), 2.18 (1H, m, H-10), 2.05 (1H, m, H-1), 1.98 (1H, m, H-2b), 1.87 (1H, m, H-8b), 1.83 (1H, m, H-2a), 1.78 (1H, m, H-8a), 1.74 (1H, m, H-9b), 1.66 (1H, m, H-9a), 1.08 (3H, m, CH3-15), 1.07 (3H, d, J 7.5 Hz, CH3-14); 13C NMR data (100 MHz, CD2Cl2): 219.3 (C-4), 177.6 (C-12), 174.2 (C-3´), 83.1 (C-6), 55.1 (C-5), 54.8 (C-2´), 52.7 (C-4´), 47.2 (C-11), 46.8 (C-1), 45.2 (C-7), 38.2 (C-1´), 36.9 (C-3), 34.8 (C-10), 33.5 (C-9), 33.3 (C-13), 25.3 (C-8), 24.3 (C-2), 16.2 (C-14), 14.3 (C-15) ; [α]D20 7.6 (c 1.00, CH2Cl2); IR spectrum (film, γ, cm-1): 2922, 1736, 1438, 1408, 1385, 1359, 1176, 1053, 1000, 910. TOFMS: 384.1844 [M+H]+ (calculated for C19H30NO5S 384.1845).
Compound 5b
1H NMR data (400 MHz, CDCl3): 4.45 (1H, d, J 5.5 Hz, H-6), δ 3.70 (3H, s, H-4´), δ 3.65 (1H, dd, J 7.1; 4.9 Hz, H-2´), δ 3.00 (1H, dd, J 13.0; 4.5 Hz, H-13b), δ 2.93 (2H, m, H-11/1´), δ 2.78 (1H, ddd, J 13.5; 7.2; 0.7 Hz, H-1´), δ 2.59 (2H, m, H-7/13a), δ 2.43 (1H, dd, J 19.2; 8.6 Hz, H-3), δ 2.21 (1H, m, H-10), δ 2.16 (1H, m, H-3a), δ 2.08 (1H, m, H-1), δ 2.03 (1H, m, H-2), δ 1.85 (1H, m, H-9b), δ 1.81 (1H, m, H-2a), δ 1.60 (1H, tdd, J 13.3; 8.7; 4.8 Hz, H-9a), δ 1.52 (2H, dd, J 9.5; 5.8 Hz, H-8a/8b), δ 1.11 (3H, s, CH3-15), 1.06 (3H, d, J 7.6 Hz, CH3-14); 13C NMR data (100 MHz, CD2Cl2): 221.7 (C-4), 176.4 (C-3´), 174.8 (C-12), 82.3 (C-6), 55.0 (C-5), 54.8 (C-2´), 52.5 (C-4´), 46.2 (C-1), 45.9(C-7), 45.8 (C-11), 38.3 (C-1´), 37.4 (C-9), 35.4 (C-10), 35.3 (C-3), 28.6 (C-13b), 24.6 (C-2), 18.4 (C-8), 16.9 (C-14), 16.1 (C-15); [α]D20 -17.5 (c 1.00, CH2Cl2); IR spectrum (film, γ, cm-1): 2921, 1831, 1732, 1620, 1435, 1408, 1385, 1340, 1176, 1053, 1000, 931, 840. TOFMS: 384.1843 [M+H]+ (calculated for C19H30NO5S 384.1845).
Acknowledgments
The authors thank the Swedish International Development Agency (SIDA) for the financial support of this study, which is part of the project “Biomolecules of medicinal and industrial interest” developed between the Universidad Mayor de San Andrés (UMSA La Paz – Bolivia) and Lund University (Sweden).
The authors thank the Swedish International Development Agency (SIDA) for the financial support of this study, which is part of the project “Biomolecules of medicinal and industrial interest” developed between the Universidad Mayor de San Andrés (UMSA La Paz – Bolivia) and Lund University (Sweden).
Conflict of interest
No author has any associations that may represent a potential conflict of interest.
No author has any associations that may represent a potential conflict of interest.
References
- Suter MB, Pagani O. (2018). Should age impact breast cancer management in young women? Fine tuning of treatment guidelines. Therapeutic advances in medical oncology 10: 1758835918776923.
- Mukohara T. (2011). Mechanisms of resistance to anti?human epidermal growth factor receptor 2 agents in breast cancer. Cancer Science 102: 1-8.
- Zhu L, Chen Y, Wei C, Yang X, Cheng J, Yang Z, Chen C, Ji Z. (2018). Anti-proliferative and pro-apoptotic effects of cinobufagin on human breast cancer MCF-7 cells and its molecular mechanism. Natural product research 32: 493-497.
- Cragg GM, Newman DJ (2013) Natural products: a continuing source of novel drug leads. Biochimica et biophysica acta 2013; 1830: 3670-3695.
- Atanasov AG, Waltenberger B, Pferschy-Wenzig EM, Linder T, Wawrosch C et al. (2015) Discovery and resupply of pharmacologically active plant-derived natural products: A review. Biotechnology advances 2015; 33: 1582-1614.
- Panda AK, Chakraborty D, Sarkar I, Khan T, Sa G. (2017). New insights into therapeutic activity and anticancer properties of curcumin. Journal of experimental pharmacology 9: 31-45.
- Weldon CB, Burow ME, Rolfe KW, Clayton JL, Jaffe BM et al. (2001). NF-κB–mediated chemoresistance in breast cancer cells. Surgery 130: 143-150.
- Zhang S, Won YK, Ong CN, Shen HM. (2005). Anti-cancer potential of sesquiterpene lactones: bioactivity and molecular mechanisms. Current medicinal chemistry. Anti-cancer agents 5: 239-249.
- Ghantous A, Gali-Muhtasib H, Vuorela H, Saliba NA, Darwiche N. (2010). What made sesquiterpene lactones reach cancer clinical trials? Drug discovery today 15: 668-678.
- Scotti MT, Fernandes MB, Ferreira MJP, Emerenciano VP. (2007). Quantitative structure–activity relationship of sesquiterpene lactones with cytotoxic activity. Bioorganic and Medicinal Chemistry 15: 2927-2934.
- Rüngeler P, Castro V, Mora G, Gören N, Vishnewski W, Pahl HL, Schmidt TJ. (1999). Inhibition of transcription factor NF-κB by sesquiterpene lactones: a proposed molecular mechanism of action. Bioorganic and Medicinal Chemistry 7: 2343-2352.
- Chen H, Chen B-Y, Liu C-T, et al. (2014). Synthesis and structure–activity relationships of guaiane-type sesquiterpene lactone derivatives with respect to inhibiting NO production in lipopolysaccharide-induced RAW 264.7 macrophages. European Journal of Medicinal Chemistry 83: 307-316.
- Garc??a-Piñeres AJ, Castro VC, Mora G, Schmidt TJ, Strunck E. (2001). Cysteine 38 in p65/NF-κB plays a crucial role in DNA binding inhibition by sesquiterpene lactones. Journal of Biological Chemistry 276: 39713-39720.
- Hehner SP, Heinrich M, Bork PM, et al. (1998). Sesquiterpene lactones specifically inhibit activation of NF-κB by preventing the degradation of IκB-α and IκB-β. Journal of Biological Chemistry 273: 1288-1297.
- Wilson AJ, Kerns JK, Callahan JF, Moody CJ. (2013). Keap calm, and carry on covalently. Journal of Medicinal Chemistry 56: 7463-7476.
- Gonzalez-Bello C. (2016). Designing Irreversible Inhibitors--Worth the Effort? ChemMedChem 11: 22-30.
- Villagomez R, Rodrigo GC, Collado IG, Calzado M, Munoz E, Åkesson B, Sterner O, Almanza G, Duan R. (2013). Multiple anticancer effects of damsin and coronopilin isolated from Ambrosia arborescens on cell cultures. Anticancer Research 33: 3799-3805.
- Villagomez R, Collado JA, Muñoz E, Almanza G, Sterner O. (2014). Natural and semi-synthetic pseudoguaianolides as inhibitors of NF-κB. Journal of Biomedical Science and Engineering 7: 833-847.
- Villagomez, R.; Quiroz, M.; Tito, A.; Sterner, O.; Almanza, G. (2015). Natural Pseudoguaianolides prepared from damsin. Chemistry of Natural Compounds51: 675-680
- Herz W, Anderson G, Gibaja S, Raulais D. (1969). Sesquiterpene lactones of some Ambrosia species. Phytochemistry 8: 877-881.
- Lozano M, Soria W, Almanza GR, Manner S, Oredsson S, Villagomez R, Sterner O. (2019). Selective cytotoxicity of damsin derivatives in breast cancer cells. Journal of Advanced Pharmaceutical Science and Technology 2: 22-36.
- Soule HD, Maloney TM, Wolman SR, Peterson WD, Brenz R, McGrath CM, Russo J, Pauley RJ, Jones RF, Brooks SC. (1990). Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Research 50: 6075-6086.
- Geltmeier A, Rinner B, Bade D, Meditz K, Witt R, Bicker U, Bludszuweit C, Maier P. (2015). Characterization of dynamic behaviour of MCF7 and MCF10A cells in ultrasonic field using modal and harmonic analyses. PLoS ONE 10: e0134999. doi:10.1371/journal.pone.0134999.
- Tanner M, Kapanen AI, Junttila T, Raheem O, Grenman S, Elo J, Elenius K, Isola J. (2004). Characterization of a novel cell line established from a patient with Herceptin-resistant breast cancer. Molecular Cancer Therapeutics 3: 1585-1592.
- Schwöbel JAH, Wondrousch D, Koleva YK, Madden JC, Cronin MTD, Schüürmann G. (2010). Prediction of Michael-type acceptor reactivity toward glutathione. Chemical Research and Toxicology 23: 1576–1585.
- Lushchak VL. (2012). Glutathione homeostasis and functions: potential targets for medical interventions. Journal of Amino Acids doi:10.1155/2012/736837.
- Schmidt TJ. (2000). Glutathione adducts of helenalin and 11α, 13-dihydrohelenalin acetate inhibit glutathione S-transferase from horse liver.Planta Medica 66: 106-109.
- Whipple RA, Vitolo MI, Boggs AE, Charpentier MS, Thompson K, Martin SS. (2013). Parthenolide and costunolide reduce microtentacles and tumor cell attachment by selectively targeting detyrosinated tubulin independent from NF-κB inhibition. Breast Cancer Research 15: R83.
- Fonrose X, Ausseil F, Soleilhac E, Masson V, David B, Pouny I, Cintrat J-C, Bernard Rousseau B, Barette C, Massiot G, and Lafanechère L. (2007) Parthenolide inhibits tubulin carboxypeptidase activity. Cancer Research 67: 2271-3378.
Citation: Maribel Lozano, Wendy Soria, Giovanna R. Almanza, Sophie Manner, Stina Oredsson, Rodrigo Villagomez and Olov Sterner. (2019). Cytotoxicity of New Damsin Derivatives in Breast Cancer Cells. Journal of Pharmacy and Drug Development 1(2). DOI: 10.5281/zenodo.3464022
Copyright: © 2019 Olov Sterner. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.